Note
This page was generated from
totalvi_in_R.ipynb.
Interactive online version:
.
Some tutorial content may look better in light mode.
CITE-seq analysis in R#
In this brief tutorial, we go over how to use scvi-tools functionality in R for analyzing CITE-seq data. We will closely follow the Bioconductor PBMC tutorial, using totalVI when appropriate.
This tutorial requires Reticulate. Please check out our installation guide for instructions on installing Reticulate and scvi-tools.
Loading and processing data with Bioconductor#
[3]:
library(BiocFileCache)
bfc <- BiocFileCache(ask=FALSE)
exprs.data <- bfcrpath(bfc, file.path(
"http://cf.10xgenomics.com/samples/cell-vdj/3.1.0",
"vdj_v1_hs_pbmc3",
"vdj_v1_hs_pbmc3_filtered_feature_bc_matrix.tar.gz"))
untar(exprs.data, exdir=tempdir())
library(DropletUtils)
sce.pbmc <- read10xCounts(file.path(tempdir(), "filtered_feature_bc_matrix"))
sce.pbmc <- splitAltExps(sce.pbmc, rowData(sce.pbmc)$Type)
Loading required package: SingleCellExperiment
Loading required package: SummarizedExperiment
Loading required package: MatrixGenerics
Loading required package: matrixStats
Attaching package: ‘MatrixGenerics’
The following objects are masked from ‘package:matrixStats’:
colAlls, colAnyNAs, colAnys, colAvgsPerRowSet, colCollapse,
colCounts, colCummaxs, colCummins, colCumprods, colCumsums,
colDiffs, colIQRDiffs, colIQRs, colLogSumExps, colMadDiffs,
colMads, colMaxs, colMeans2, colMedians, colMins, colOrderStats,
colProds, colQuantiles, colRanges, colRanks, colSdDiffs, colSds,
colSums2, colTabulates, colVarDiffs, colVars, colWeightedMads,
colWeightedMeans, colWeightedMedians, colWeightedSds,
colWeightedVars, rowAlls, rowAnyNAs, rowAnys, rowAvgsPerColSet,
rowCollapse, rowCounts, rowCummaxs, rowCummins, rowCumprods,
rowCumsums, rowDiffs, rowIQRDiffs, rowIQRs, rowLogSumExps,
rowMadDiffs, rowMads, rowMaxs, rowMeans2, rowMedians, rowMins,
rowOrderStats, rowProds, rowQuantiles, rowRanges, rowRanks,
rowSdDiffs, rowSds, rowSums2, rowTabulates, rowVarDiffs, rowVars,
rowWeightedMads, rowWeightedMeans, rowWeightedMedians,
rowWeightedSds, rowWeightedVars
Loading required package: GenomicRanges
Loading required package: stats4
Loading required package: BiocGenerics
Attaching package: ‘BiocGenerics’
The following objects are masked from ‘package:stats’:
IQR, mad, sd, var, xtabs
The following objects are masked from ‘package:base’:
anyDuplicated, append, as.data.frame, basename, cbind, colnames,
dirname, do.call, duplicated, eval, evalq, Filter, Find, get, grep,
grepl, intersect, is.unsorted, lapply, Map, mapply, match, mget,
order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank,
rbind, Reduce, rownames, sapply, setdiff, sort, table, tapply,
union, unique, unsplit, which.max, which.min
Loading required package: S4Vectors
Attaching package: ‘S4Vectors’
The following objects are masked from ‘package:base’:
expand.grid, I, unname
Loading required package: IRanges
Loading required package: GenomeInfoDb
Loading required package: Biobase
Welcome to Bioconductor
Vignettes contain introductory material; view with
'browseVignettes()'. To cite Bioconductor, see
'citation("Biobase")', and for packages 'citation("pkgname")'.
Attaching package: ‘Biobase’
The following object is masked from ‘package:MatrixGenerics’:
rowMedians
The following objects are masked from ‘package:matrixStats’:
anyMissing, rowMedians
Pre-processing and quality control#
[4]:
unfiltered <- sce.pbmc
[5]:
library(scater)
is.mito <- grep("^MT-", rowData(sce.pbmc)$Symbol)
stats <- perCellQCMetrics(sce.pbmc, subsets=list(Mito=is.mito))
high.mito <- isOutlier(stats$subsets_Mito_percent, type="higher")
low.adt <- stats$`altexps_Antibody Capture_detected` < nrow(altExp(sce.pbmc))/2
discard <- high.mito | low.adt
sce.pbmc <- sce.pbmc[,!discard]
Loading required package: scuttle
Loading required package: ggplot2
[6]:
summary(high.mito)
Mode FALSE TRUE
logical 6660 571
[7]:
colData(unfiltered) <- cbind(colData(unfiltered), stats)
unfiltered$discard <- discard
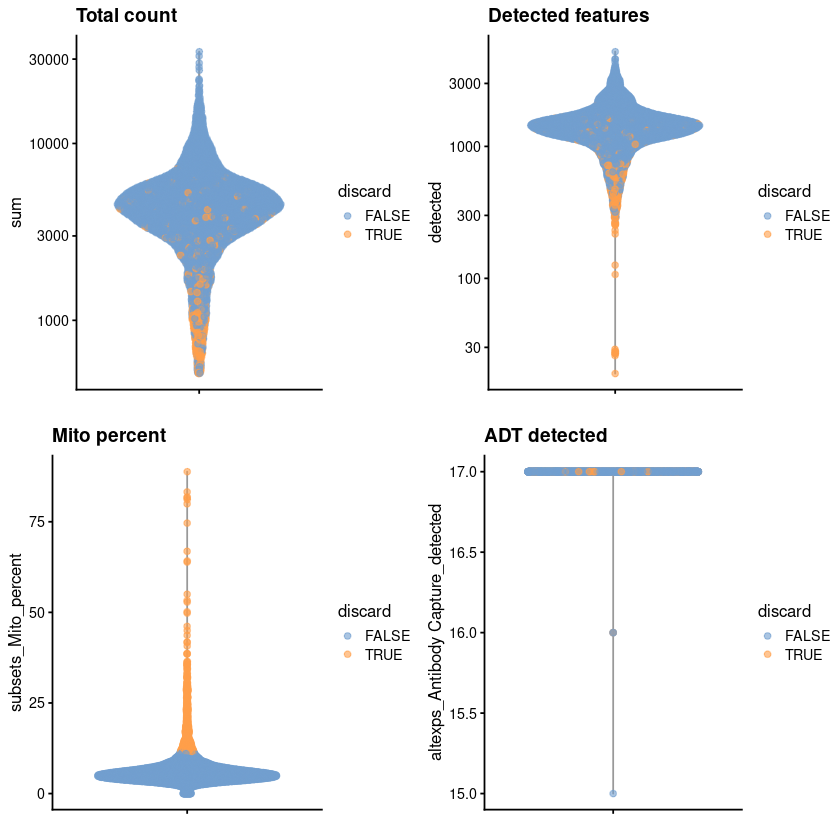
gridExtra::grid.arrange(
plotColData(unfiltered, y="sum", colour_by="discard") +
scale_y_log10() + ggtitle("Total count"),
plotColData(unfiltered, y="detected", colour_by="discard") +
scale_y_log10() + ggtitle("Detected features"),
plotColData(unfiltered, y="subsets_Mito_percent",
colour_by="discard") + ggtitle("Mito percent"),
plotColData(unfiltered, y="altexps_Antibody Capture_detected",
colour_by="discard") + ggtitle("ADT detected"),
ncol=2
)
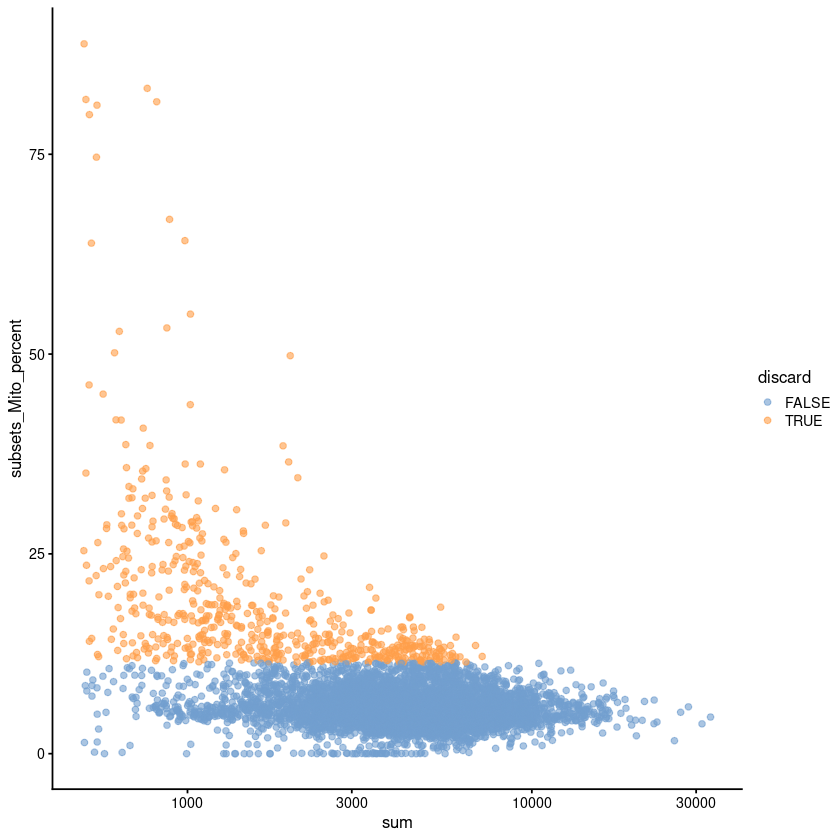
[8]:
plotColData(unfiltered, x="sum", y="subsets_Mito_percent",
colour_by="discard") + scale_x_log10()
Normalization#
While we normalize the data here using standard Bioconductor practices, we will use the counts later for totalVI.
[11]:
library(scran)
set.seed(1000)
clusters <- quickCluster(sce.pbmc)
sce.pbmc <- computeSumFactors(sce.pbmc, cluster=clusters)
altExp(sce.pbmc) <- computeMedianFactors(altExp(sce.pbmc))
sce.pbmc <- logNormCounts(sce.pbmc, use_altexps=TRUE)
Warning message:
“'use.altexps=' is deprecated.
Use 'applySCE(x, logNormCounts)' instead.”
Data conversion (SCE -> AnnData)#
We use sceasy for conversion, and load the necessary Python packages for later.
[30]:
library(reticulate)
library(sceasy)
library(anndata)
use_python("/home/adam/.pyenv/versions/3.9.7/bin/python", required = TRUE)
sc <- import("scanpy", convert = FALSE)
scvi <- import("scvi", convert = FALSE)
sys <- import ("sys", convert = FALSE)
We make two AnnData objects, one per modality, and then store the protein counts in the canonical location for scvi-tools.
[24]:
adata <- convertFormat(sce.pbmc, from="sce", to="anndata", main_layer="counts", drop_single_values=FALSE)
adata_protein <- convertFormat(altExp(sce.pbmc), from="sce", to="anndata", main_layer="counts", drop_single_values=FALSE)
adata$obsm["protein"] <- adata_protein$to_df()
[28]:
adata
AnnData object with n_obs × n_vars = 6660 × 33538
obs: 'Sample', 'Barcode', 'sizeFactor'
var: 'ID', 'Symbol', 'Type'
obsm: 'protein'
Run totalVI for dimensionality reduction#
totalVI will output a low-dimensional representation of cells that captures information from both the RNA and protein. Here we show how to use totalVI for only dimensionality reduction, though totalVI can perform other tasks that are shown in the Python-based tutorials. The intention here is to provide some examples of how to use totalVI from R.
[31]:
scvi$model$TOTALVI$setup_anndata(adata, protein_expression_obsm_key="protein")
None
None
[32]:
vae <- scvi$model$TOTALVI(adata)
vae$train()
None
[38]:
reducedDims(sce.pbmc) <- list(TOTALVI=py_to_r(vae$get_latent_representation()))
sce.pbmc <- runUMAP(sce.pbmc, dimred="TOTALVI")
sce.pbmc <- runTSNE(sce.pbmc, dimred="TOTALVI")
Clustering#
[39]:
g <- buildSNNGraph(sce.pbmc, k=10, use.dimred = 'TOTALVI')
clust <- igraph::cluster_walktrap(g)$membership
colLabels(sce.pbmc) <- factor(clust)
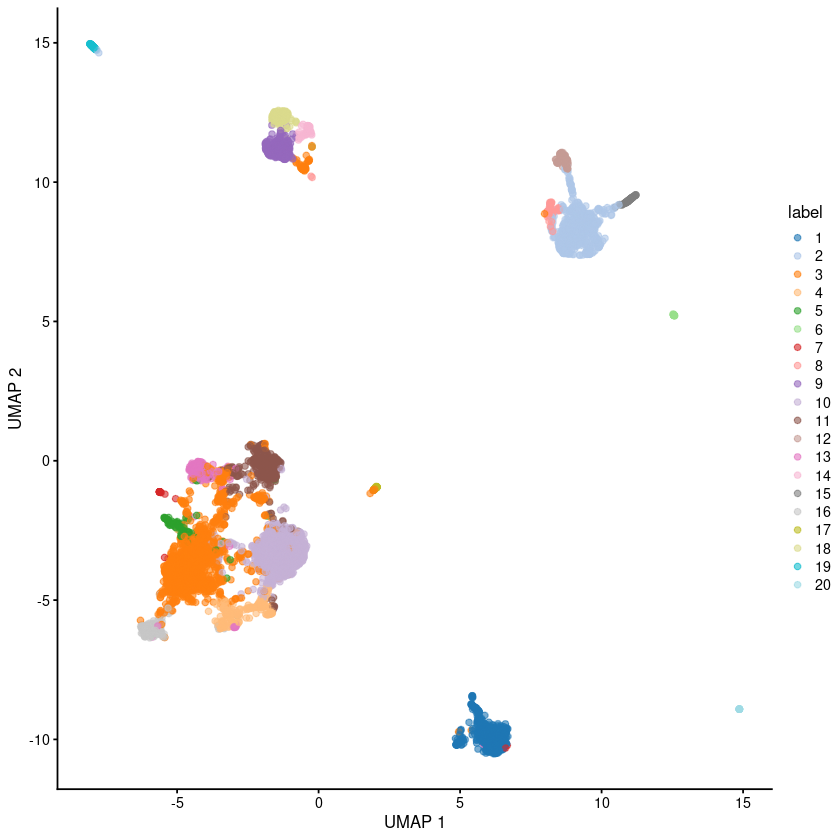
[37]:
plotUMAP(sce.pbmc, colour_by="label")
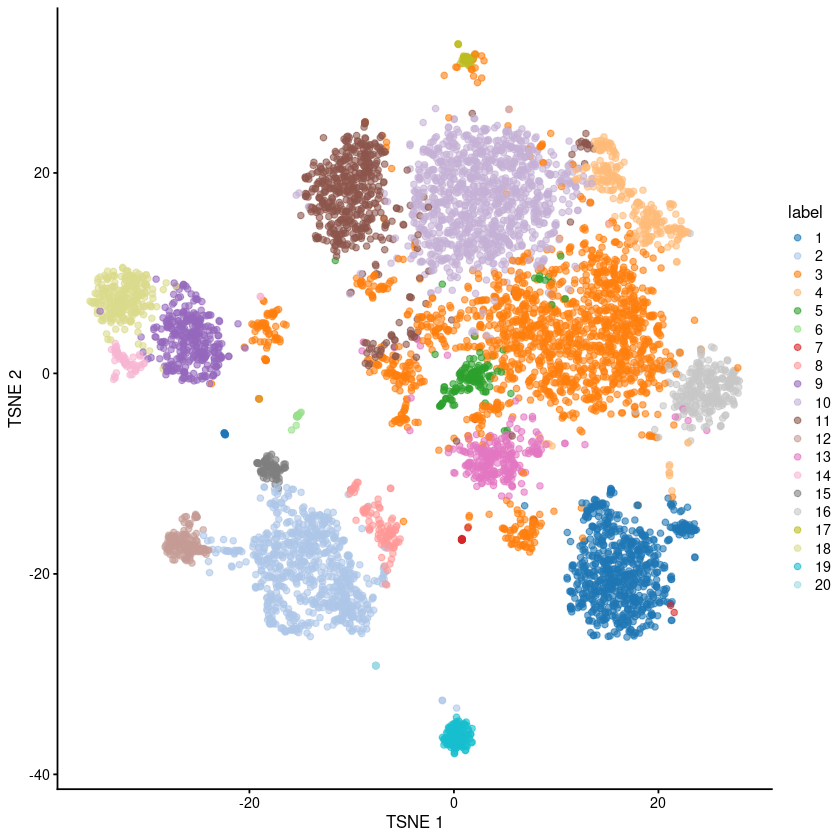
[40]:
plotTSNE(sce.pbmc, colour_by="label")
[43]:
sI <- sessionInfo()
sI$loadedOnly <- NULL
print(sI, locale=FALSE)
R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 20.04.2 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] anndata_0.7.5.3 sceasy_0.0.6
[3] reticulate_1.20 scran_1.21.3
[5] scater_1.21.3 ggplot2_3.3.5
[7] scuttle_1.3.1 DropletUtils_1.13.2
[9] SingleCellExperiment_1.15.2 SummarizedExperiment_1.23.4
[11] Biobase_2.53.0 GenomicRanges_1.45.0
[13] GenomeInfoDb_1.29.8 IRanges_2.27.2
[15] S4Vectors_0.31.3 BiocGenerics_0.39.2
[17] MatrixGenerics_1.5.4 matrixStats_0.60.1
[19] BiocFileCache_2.1.1 dbplyr_2.1.1