10. Advanced autotune tutorial¶
DISCLAIMER: Most experiments in this notebook require one or more GPUs to keep their runtime a matter of hours. DISCLAIMER: To use our new autotune feature in parallel mode, you need to install `MongoDb <https://docs.mongodb.com/manual/installation/>`__ first.
In this notebook, we give an in-depth tutorial on scVI’s new autotune module.
Overall, the new module enables users to perform parallel hyperparemter search for any scVI model and on any number of GPUs/CPUs. Although, the search may be performed sequentially using only one GPU/CPU, we will focus on the paralel case. Note that GPUs provide a much faster approach as they are particularly suitable for neural networks gradient back-propagation.
Additionally, we provide the code used to generate the results presented in our Hyperoptimization blog post. For an in-depth analysis of the results obtained on three gold standard scRNAseq datasets (Cortex, PBMC and BrainLarge), please to the above blog post. In the blog post, we also suggest guidelines on how and when to use our auto-tuning feature.
[1]:
import sys
sys.path.append("../../")
sys.path.append("../")
%matplotlib inline
/home/ec2-user
[2]:
import logging
import os
import pickle
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import torch
from hyperopt import hp
from scvi.dataset import BrainLargeDataset, CortexDataset, LoomDataset, PbmcDataset
from scvi.inference import UnsupervisedTrainer
from scvi.inference.autotune import auto_tune_scvi_model
from scvi.models import VAE
[ ]:
logger = logging.getLogger("scvi.inference.autotune")
logger.setLevel(logging.WARNING)
[2]:
def allow_notebook_for_test():
print("Testing the autotune advanced notebook")
test_mode = False
def if_not_test_else(x, y):
if not test_mode:
return x
else:
return y
save_path = "data/"
n_epochs = if_not_test_else(1000, 1)
n_epochs_brain_large = if_not_test_else(50, 1)
max_evals = if_not_test_else(100, 1)
reserve_timeout = if_not_test_else(180, 5)
fmin_timeout = if_not_test_else(300, 10)
10.1. Default usage¶
For the sake of principled simplicity, we provide an all-default approach to hyperparameter search for any scVI model. The few lines below present an example of how to perform hyper-parameter search for scVI on the Cortex dataset.
Note that, by default, the model used is scVI’s VAE and the trainer is the UnsupervisedTrainer
Also, the default search space is as follows: * n_latent: [5, 15] * n_hidden: {64, 128, 256} * n_layers: [1, 5] * dropout_rate: {0.1, 0.3, 0.5, 0.7} * reconstruction_loss: {“zinb”, “nb”} * lr: {0.01, 0.005, 0.001, 0.0005, 0.0001}
On a more practical note, verbosity varies in the following way: * logger.setLevel(logging.WARNING) will show a progress bar. * logger.setLevel(logging.INFO) will show global logs including the number of jobs done. * logger.setLevel(logging.DEBUG) will show detailed logs for each training (e.g the parameters tested).
This function’s behaviour can be customized, please refer to the rest of this tutorial as well as its documentation for information about the different parameters available.
10.1.1. Running the hyperoptimization process.¶
[ ]:
cortex_dataset = CortexDataset(save_path=save_path)
INFO:scvi.dataset.dataset:File tests/data/expression.bin already downloaded
INFO:scvi.dataset.cortex:Loading Cortex data
[5]:
best_trainer, trials = auto_tune_scvi_model(
gene_dataset=cortex_dataset,
parallel=True,
exp_key="cortex_dataset",
train_func_specific_kwargs={"n_epochs": n_epochs},
max_evals=max_evals,
reserve_timeout=reserve_timeout,
fmin_timeout=fmin_timeout,
)
100%|███████| 100/100 [3:21:15<00:00, 102.45s/it]
10.1.2. Returned objects¶
The trials object contains detailed information about each run. trials.trials is an Iterable in which each element corresponds to a single run. It can be used as a dictionary for wich the key “result” yields a dictionnary containing the outcome of the run as defined in our default objective function (or the user’s custom version). For example, it will contain information on the hyperparameters used (under the “space” key), the resulting metric (under the “loss” key) or the status of
the run.
The best_trainer object can be used directly as an scVI Trainer object. It is the result of a training on the whole dataset provided using the optimal set of hyperparameters found.
10.2. Custom hyperamater space¶
Although our default can be a good one in a number of cases, we still provide an easy way to use custom values for the hyperparameters search space. These are broken down in three categories: * Hyperparameters for the Trainer instance. (if any) * Hyperparameters for the Trainer instance’s train method. (e.g lr) * Hyperparameters for the model instance. (e.g n_layers)
To build your own hyperparameter space follow the scheme used in scVI’s codebase as well as the sample below. Note the various spaces you define, have to follow the hyperopt syntax, for which you can find a detailed description here.
For example, if you were to want to search over a continuous range of droupouts varying in [0.1, 0.3] and for a continuous learning rate varying in [0.001, 0.0001], you could use the following search space.
[ ]:
space = {
"model_tunable_kwargs": {"dropout_rate": hp.uniform("dropout_rate", 0.1, 0.3)},
"train_func_tunable_kwargs": {"lr": hp.loguniform("lr", -4.0, -3.0)},
}
best_trainer, trials = auto_tune_scvi_model(
gene_dataset=cortex_dataset,
space=space,
parallel=True,
exp_key="cortex_dataset_custom_space",
train_func_specific_kwargs={"n_epochs": n_epochs},
max_evals=max_evals,
reserve_timeout=reserve_timeout,
fmin_timeout=fmin_timeout,
)
10.3. Custom objective metric¶
By default, our autotune process tracks the marginal negative log likelihood of the best state of the model according ot the held-out Evidence Lower BOund (ELBO). But, if you want to track a different early stopping metric and optimize a different loss you can use auto_tune_scvi_model’s parameters.
For example, if for some reason, you had a dataset coming from two batches (i.e two merged datasets) and wanted to optimize the hyperparameters for the batch mixing entropy. You could use the code below, which makes use of the metric_name argument of auto_tune_scvi_model. This can work for any metric that is implemented in the Posterior class you use. You may also specify the name of the Posterior attribute you want to use (e.g “train_set”).
[ ]:
pbmc_dataset = PbmcDataset(
save_path=save_path, save_path_10X=os.path.join(save_path, "10X")
)
[ ]:
best_trainer, trials = auto_tune_scvi_model(
gene_dataset=pbmc_dataset,
metric_name="entropy_batch_mixing",
posterior_name="train_set",
parallel=True,
exp_key="pbmc_entropy_batch_mixing",
train_func_specific_kwargs={"n_epochs": n_epochs},
max_evals=max_evals,
reserve_timeout=reserve_timeout,
fmin_timeout=fmin_timeout,
)
10.4. Custom objective function¶
Below, we describe, using one of our Synthetic dataset, how to tune our annotation model SCANVI for, e.g, better accuracy on a 20% subset of the labelled data. Note that the model is trained in a semi-supervised framework, that is why we have a labelled and unlabelled dataset. Please, refer to the original paper for details on SCANVI!
In this case, as described in our annotation notebook we may want to form the labelled/unlabelled sets using batch indices. Unfortunately, that requires a little “by hand” work. Even in that case, we are able to leverage the new autotune module to perform hyperparameter tuning. In order to do so, one has to write his own objective function and feed it to auto_tune_scvi_model.
One can proceed as described below. Note three important conditions: * Since it is going to be pickled the objective should not be implemented in the “main” module, i.e an executable script or a notebook. * the objective should have the search space as its first attribute and a boolean is_best_training as its second. * If not using a cutstom search space, it should be expected to take the form of a dictionary with the following keys: * "model_tunable_kwargs" *
"trainer_tunable_kwargs" * "train_func_tunable_kwargs"
[ ]:
synthetic_dataset = LoomDataset(
filename="simulation_1.loom",
save_path=os.path.join(save_path, "simulation/"),
url="https://github.com/YosefLab/scVI-data/raw/master/simulation/simulation_1.loom",
)
[ ]:
from notebooks.utils.autotune_advanced_notebook import custom_objective_hyperopt
[ ]:
objective_kwargs = dict(dataset=synthetic_dataset, n_epochs=n_epochs)
best_trainer, trials = auto_tune_scvi_model(
custom_objective_hyperopt=custom_objective_hyperopt,
objective_kwargs=objective_kwargs,
parallel=True,
exp_key="synthetic_dataset_scanvi",
max_evals=max_evals,
reserve_timeout=reserve_timeout,
fmin_timeout=fmin_timeout,
)
10.5. Delayed populating, for very large datasets.¶
DISCLAIMER: We don’t actually need this for the BrainLarge dataset with 720 genes, this is just an example.
The fact is that after building the objective function and feeding it to hyperopt, it is pickled on to the MongoWorkers. Thus, if you pass a loaded dataset as a partial argument to the objective function, and this dataset exceeds 4Gb, you’ll get a PickleError (Objects larger than 4Gb can’t be pickled).
To remedy this issue, in case you have a very large dataset for which you want to perform hyperparameter optimization, you should subclass scVI’s DownloadableDataset or use one of its many existing subclasses, such that the dataset can be populated inside the objective function which is called by each worker.
[ ]:
brain_large_dataset = BrainLargeDataset(save_path=save_path, delayed_populating=True)
best_trainer, trials = auto_tune_scvi_model(
gene_dataset=brain_large_dataset,
delayed_populating=True,
parallel=True,
exp_key="brain_large_dataset",
max_evals=max_evals,
trainer_specific_kwargs={
"early_stopping_kwargs": {
"early_stopping_metric": "elbo",
"save_best_state_metric": "elbo",
"patience": 20,
"threshold": 0,
"reduce_lr_on_plateau": True,
"lr_patience": 10,
"lr_factor": 0.2,
}
},
train_func_specific_kwargs={"n_epochs": n_epochs_brain_large},
reserve_timeout=reserve_timeout,
fmin_timeout=fmin_timeout,
)
10.6. Blog post reproducibility¶
Below, we provide some code to reproduce the results of our blog post on hyperparameter search with scVI. Note, that this can also be used as a tutorial on how to make senss of the output of the autotuning process, the Trials object.
10.7. Cortex, Pbmc and BrainLarge hyperparameter optimization¶
First off, we run the default hyperparameter optimization procedure (default search space, 100 runs) on each of the three dataset of our study: * The Cortex dataset (done above) * The Pbmc dataset * The Brain Large dataset (done above)
[ ]:
best_trainer, trials = auto_tune_scvi_model(
gene_dataset=pbmc_dataset,
parallel=True,
exp_key="pbmc_dataset",
max_evals=max_evals,
train_func_specific_kwargs={"n_epochs": n_epochs},
reserve_timeout=reserve_timeout,
fmin_timeout=fmin_timeout,
)
INFO:scvi.inference.autotune:Starting experiment: pbmc_bis
DEBUG:scvi.inference.autotune:Using default parameter search space.
INFO:scvi.inference.autotune:Fixed parameters:
model:
{}
trainer:
{'early_stopping_kwargs': {'early_stopping_metric': 'll', 'save_best_state_metric': 'll', 'patience': 50, 'threshold': 0, 'reduce_lr_on_plateau': False, 'lr_patience': 25, 'lr_factor': 0.2}}
train method:
{}
INFO:scvi.inference.autotune:Starting parallel hyperoptimization
DEBUG:scvi.inference.autotune:Starting MongoDb process, logs redirected to ./mongo/mongo_logfile.txt.
DEBUG:scvi.inference.autotune:Starting minimization procedure
DEBUG:scvi.inference.autotune:Starting worker launcher
DEBUG:scvi.inference.autotune:Instantiating trials object.
DEBUG:scvi.inference.autotune:Started waiting for fmin Process.
DEBUG:scvi.inference.autotune:Started waiting for Worker Launcher.
DEBUG:scvi.inference.autotune:gpu_ids is None, defaulting to all {n_gpus} GPUs found by torch.
DEBUG:scvi.inference.autotune:Calling fmin.
INFO:scvi.inference.autotune:Starting 1 worker.s for each of the 1 gpu.s set for use/found.
INFO:scvi.inference.autotune:Starting 0 cpu worker.s
DEBUG:scvi.inference.autotune:Worker working...
INFO:hyperopt.mongoexp:PROTOCOL mongo
INFO:hyperopt.mongoexp:USERNAME None
INFO:hyperopt.mongoexp:HOSTNAME localhost
INFO:hyperopt.mongoexp:PORT 1234
INFO:hyperopt.mongoexp:PATH /scvi_db/jobs
INFO:hyperopt.mongoexp:AUTH DB None
INFO:hyperopt.mongoexp:DB scvi_db
INFO:hyperopt.mongoexp:COLLECTION jobs
DEBUG:hyperopt.mongoexp:job found: SON([('_id', ObjectId('5ce40640cd2327ac67fdbd3e')), ('state', 1), ('tid', 1176), ('spec', None), ('result', SON([('status', 'new')])), ('misc', SON([('tid', 1176), ('cmd', ['domain_attachment', 'FMinIter_Domain']), ('workdir', None), ('idxs', SON([('dropout_rate', [1176]), ('lr', [1176]), ('n_hidden', [1176]), ('n_latent', [1176]), ('n_layers', [1176]), ('reconstruction_loss', [1176])])), ('vals', SON([('dropout_rate', [0]), ('lr', [1]), ('n_hidden', [0]), ('n_latent', [4]), ('n_layers', [4]), ('reconstruction_loss', [0])]))])), ('exp_key', 'pbmc_bis'), ('owner', ['ip-172-31-3-254:19748']), ('version', 0), ('book_time', datetime.datetime(2019, 5, 21, 14, 8, 1, 246000)), ('refresh_time', datetime.datetime(2019, 5, 21, 14, 8, 1, 246000))])
DEBUG:scvi.inference.autotune:Listener listening...
INFO:scvi.inference.autotune:Parameters being tested:
model:
{'dropout_rate': 0.1, 'n_hidden': 64, 'n_latent': 9, 'n_layers': 5, 'reconstruction_loss': 'zinb'}
trainer:
{'early_stopping_kwargs': {'early_stopping_metric': 'll', 'save_best_state_metric': 'll', 'patience': 50, 'threshold': 0, 'reduce_lr_on_plateau': False, 'lr_patience': 25, 'lr_factor': 0.2}, 'use_cuda': True, 'show_progbar': False, 'frequency': 1}
train method:
{'lr': 0.005, 'n_epochs': 1000}
DEBUG:scvi.inference.autotune:Instantiating model
DEBUG:scvi.inference.autotune:Instantiating trainer
DEBUG:scvi.inference.autotune:Starting training
DEBUG:scvi.inference.autotune:Finished training
DEBUG:scvi.inference.autotune:Training of 256 epochs finished in 0:11:10.437926 with loss = 1323.5555671392826
INFO:hyperopt.mongoexp:job finished: 5ce40640cd2327ac67fdbd3e
INFO:scvi.inference.autotune:1 job.s done
INFO:progress_logger:None
DEBUG:hyperopt.mongoexp:job found: SON([('_id', ObjectId('5ce40641cd2327ac67fdbd3f')), ('state', 1), ('tid', 1177), ('spec', None), ('result', SON([('status', 'new')])), ('misc', SON([('tid', 1177), ('cmd', ['domain_attachment', 'FMinIter_Domain']), ('workdir', None), ('idxs', SON([('dropout_rate', [1177]), ('lr', [1177]), ('n_hidden', [1177]), ('n_latent', [1177]), ('n_layers', [1177]), ('reconstruction_loss', [1177])])), ('vals', SON([('dropout_rate', [4]), ('lr', [4]), ('n_hidden', [1]), ('n_latent', [4]), ('n_layers', [2]), ('reconstruction_loss', [1])]))])), ('exp_key', 'pbmc_bis'), ('owner', ['ip-172-31-3-254:19748']), ('version', 0), ('book_time', datetime.datetime(2019, 5, 21, 14, 19, 11, 962000)), ('refresh_time', datetime.datetime(2019, 5, 21, 14, 19, 11, 962000))])
INFO:scvi.inference.autotune:Parameters being tested:
model:
{'dropout_rate': 0.9, 'n_hidden': 128, 'n_latent': 9, 'n_layers': 3, 'reconstruction_loss': 'nb'}
trainer:
{'early_stopping_kwargs': {'early_stopping_metric': 'll', 'save_best_state_metric': 'll', 'patience': 50, 'threshold': 0, 'reduce_lr_on_plateau': False, 'lr_patience': 25, 'lr_factor': 0.2}, 'use_cuda': True, 'show_progbar': False, 'frequency': 1}
train method:
{'lr': 0.0001, 'n_epochs': 1000}
DEBUG:scvi.inference.autotune:Instantiating model
DEBUG:scvi.inference.autotune:Instantiating trainer
DEBUG:scvi.inference.autotune:Starting training
DEBUG:scvi.inference.autotune:Finished training
DEBUG:scvi.inference.autotune:Training of 1002 epochs finished in 0:35:13.787563 with loss = 1374.7283445840283
INFO:hyperopt.mongoexp:job finished: 5ce40641cd2327ac67fdbd3f
INFO:scvi.inference.autotune:2 job.s done
INFO:progress_logger:None
DEBUG:hyperopt.mongoexp:job found: SON([('_id', ObjectId('5ce408e0cd2327ac67fdbd40')), ('state', 1), ('tid', 1178), ('spec', None), ('result', SON([('status', 'new')])), ('misc', SON([('tid', 1178), ('cmd', ['domain_attachment', 'FMinIter_Domain']), ('workdir', None), ('idxs', SON([('dropout_rate', [1178]), ('lr', [1178]), ('n_hidden', [1178]), ('n_latent', [1178]), ('n_layers', [1178]), ('reconstruction_loss', [1178])])), ('vals', SON([('dropout_rate', [0]), ('lr', [3]), ('n_hidden', [0]), ('n_latent', [3]), ('n_layers', [3]), ('reconstruction_loss', [0])]))])), ('exp_key', 'pbmc_bis'), ('owner', ['ip-172-31-3-254:19748']), ('version', 0), ('book_time', datetime.datetime(2019, 5, 21, 14, 54, 26, 16000)), ('refresh_time', datetime.datetime(2019, 5, 21, 14, 54, 26, 16000))])
INFO:scvi.inference.autotune:Parameters being tested:
model:
{'dropout_rate': 0.1, 'n_hidden': 64, 'n_latent': 8, 'n_layers': 4, 'reconstruction_loss': 'zinb'}
trainer:
{'early_stopping_kwargs': {'early_stopping_metric': 'll', 'save_best_state_metric': 'll', 'patience': 50, 'threshold': 0, 'reduce_lr_on_plateau': False, 'lr_patience': 25, 'lr_factor': 0.2}, 'use_cuda': True, 'show_progbar': False, 'frequency': 1}
train method:
{'lr': 0.0005, 'n_epochs': 1000}
DEBUG:scvi.inference.autotune:Instantiating model
DEBUG:scvi.inference.autotune:Instantiating trainer
DEBUG:scvi.inference.autotune:Starting training
DEBUG:scvi.inference.autotune:Finished training
DEBUG:scvi.inference.autotune:Training of 681 epochs finished in 0:27:37.825918 with loss = 1323.6787068650958
INFO:hyperopt.mongoexp:job finished: 5ce408e0cd2327ac67fdbd40
INFO:scvi.inference.autotune:3 job.s done
INFO:progress_logger:None
DEBUG:hyperopt.mongoexp:job found: SON([('_id', ObjectId('5ce41122cd2327ac67fdbd41')), ('state', 1), ('tid', 1179), ('spec', None), ('result', SON([('status', 'new')])), ('misc', SON([('tid', 1179), ('cmd', ['domain_attachment', 'FMinIter_Domain']), ('workdir', None), ('idxs', SON([('dropout_rate', [1179]), ('lr', [1179]), ('n_hidden', [1179]), ('n_latent', [1179]), ('n_layers', [1179]), ('reconstruction_loss', [1179])])), ('vals', SON([('dropout_rate', [2]), ('lr', [1]), ('n_hidden', [2]), ('n_latent', [2]), ('n_layers', [4]), ('reconstruction_loss', [1])]))])), ('exp_key', 'pbmc_bis'), ('owner', ['ip-172-31-3-254:19748']), ('version', 0), ('book_time', datetime.datetime(2019, 5, 21, 15, 22, 4, 77000)), ('refresh_time', datetime.datetime(2019, 5, 21, 15, 22, 4, 77000))])
INFO:scvi.inference.autotune:Parameters being tested:
model:
{'dropout_rate': 0.5, 'n_hidden': 256, 'n_latent': 7, 'n_layers': 5, 'reconstruction_loss': 'nb'}
trainer:
{'early_stopping_kwargs': {'early_stopping_metric': 'll', 'save_best_state_metric': 'll', 'patience': 50, 'threshold': 0, 'reduce_lr_on_plateau': False, 'lr_patience': 25, 'lr_factor': 0.2}, 'use_cuda': True, 'show_progbar': False, 'frequency': 1}
train method:
{'lr': 0.005, 'n_epochs': 1000}
DEBUG:scvi.inference.autotune:Instantiating model
DEBUG:scvi.inference.autotune:Instantiating trainer
DEBUG:scvi.inference.autotune:Starting training
DEBUG:scvi.inference.autotune:Finished training
DEBUG:scvi.inference.autotune:Training of 240 epochs finished in 0:09:53.742285 with loss = 1326.2741477272727
INFO:hyperopt.mongoexp:job finished: 5ce41122cd2327ac67fdbd41
INFO:scvi.inference.autotune:4 job.s done
INFO:progress_logger:None
DEBUG:hyperopt.mongoexp:job found: SON([('_id', ObjectId('5ce4179ccd2327ac67fdbd42')), ('state', 1), ('tid', 1180), ('spec', None), ('result', SON([('status', 'new')])), ('misc', SON([('tid', 1180), ('cmd', ['domain_attachment', 'FMinIter_Domain']), ('workdir', None), ('idxs', SON([('dropout_rate', [1180]), ('lr', [1180]), ('n_hidden', [1180]), ('n_latent', [1180]), ('n_layers', [1180]), ('reconstruction_loss', [1180])])), ('vals', SON([('dropout_rate', [2]), ('lr', [3]), ('n_hidden', [1]), ('n_latent', [9]), ('n_layers', [1]), ('reconstruction_loss', [0])]))])), ('exp_key', 'pbmc_bis'), ('owner', ['ip-172-31-3-254:19748']), ('version', 0), ('book_time', datetime.datetime(2019, 5, 21, 15, 31, 58, 54000)), ('refresh_time', datetime.datetime(2019, 5, 21, 15, 31, 58, 54000))])
INFO:scvi.inference.autotune:Parameters being tested:
model:
{'dropout_rate': 0.5, 'n_hidden': 128, 'n_latent': 14, 'n_layers': 2, 'reconstruction_loss': 'zinb'}
trainer:
{'early_stopping_kwargs': {'early_stopping_metric': 'll', 'save_best_state_metric': 'll', 'patience': 50, 'threshold': 0, 'reduce_lr_on_plateau': False, 'lr_patience': 25, 'lr_factor': 0.2}, 'use_cuda': True, 'show_progbar': False, 'frequency': 1}
train method:
{'lr': 0.0005, 'n_epochs': 1000}
DEBUG:scvi.inference.autotune:Instantiating model
DEBUG:scvi.inference.autotune:Instantiating trainer
DEBUG:scvi.inference.autotune:Starting training
10.8. Handy class to handle the results of each experiment¶
In the helper, autotune_advanced_notebook.py we have implemented a Benchmarkable class which will help with things such as benchmark computation, results visualisation in dataframes, etc.
[ ]:
from notebooks.utils.autotune_advanced_notebook import Benchmarkable
10.9. Make experiment benchmarkables¶
Below, we use our helper class to store and process the results of the experiments. It allows us to generate: * Imputed values from scVI * Dataframes containing: * For each dataset, the results of each trial along with the parameters used. * For all dataset, the best result and the associated hyperparameters
[2]:
results_path = "."
10.9.1. Compute imputed values¶
[ ]:
cortex = Benchmarkable(
global_path=results_path, exp_key="cortex_dataset", name="Cortex tuned"
)
cortex.compute_imputed()
pbmc = Benchmarkable(
global_path=results_path, exp_key="pbmc_dataset", name="Pbmc tuned"
)
pbmc.compute_imputed()
brain_large = Benchmarkable(
global_path=results_path, exp_key="brain_large_dataset", name="Brain Large tuned"
)
brain_large.compute_imputed()
training: 100%|██████████| 248/248 [01:04<00:00, 3.90it/s]
Median of Median: 2.0815
Mean of Median for each cell: 2.8750
training: 100%|██████████| 160/160 [03:10<00:00, 1.19s/it]
Median of Median: 0.8515
Mean of Median for each cell: 0.9372
training: 100%|██████████| 170/170 [03:40<00:00, 1.29s/it]
Median of Median: 0.8394
Mean of Median for each cell: 0.9246
training: 88%|████████▊ | 44/50 [1:24:35<11:31, 115.28s/it]
10.10. Compute results with default parameters¶
Below we compute the results obtained with default hyperparameters for each dataset in the study.
10.10.1. Train each VAE¶
[ ]:
n_epochs_one_shot = if_not_test_else(400, 1)
[8]:
vae = VAE(cortex_dataset.nb_genes, n_batch=cortex_dataset.n_batches * False)
trainer = UnsupervisedTrainer(
vae, cortex_dataset, train_size=0.75, use_cuda=True, frequency=1
)
trainer.train(n_epochs=n_epochs_one_shot, lr=1e-3)
with open("trainer_cortex_one_shot", "wb") as f:
pickle.dump(trainer, f)
with open("model_cortex_one_shot", "wb") as f:
torch.save(vae, f)
training: 100%|██████████| 400/400 [02:31<00:00, 2.63it/s]
[9]:
vae = VAE(pbmc_dataset.nb_genes, n_batch=pbmc_dataset.n_batches * False)
trainer = UnsupervisedTrainer(
vae, pbmc_dataset, train_size=0.75, use_cuda=True, frequency=1
)
trainer.train(n_epochs=n_epochs_one_shot, lr=1e-3)
with open("trainer_pbmc_one_shot", "wb") as f:
pickle.dump(trainer, f)
with open("model_pbmc_one_shot", "wb") as f:
torch.save(vae, f)
training: 100%|██████████| 400/400 [15:54<00:00, 2.39s/it]
[10]:
vae = VAE(brain_large_dataset.nb_genes, n_batch=brain_large_dataset.n_batches * False)
trainer = UnsupervisedTrainer(
vae, brain_large_dataset, train_size=0.75, use_cuda=True, frequency=1
)
trainer.train(n_epochs=n_epochs_brain_large, lr=1e-3)
with open("trainer_brain_large_one_shot", "wb") as f:
pickle.dump(trainer, f)
with open("model_brain_large_one_shot", "wb") as f:
torch.save(vae, f)
training: 100%|██████████| 50/50 [2:28:23<00:00, 178.25s/it]
Again, we use our helper class to contain, preprocess and access the results of each experiment.
[11]:
cortex_one_shot = Benchmarkable(
trainer_fname="trainer_cortex_one_shot",
model_fname="model_cortex_one_shot",
name="Cortex default",
is_one_shot=True,
)
cortex_one_shot.compute_imputed(n_epochs=n_epochs_one_shot)
pbmc_one_shot = Benchmarkable(
trainer_fname="trainer_pbmc_one_shot",
model_fname="model_pbmc_one_shot",
name="Pbmc default",
is_one_shot=True,
)
pbmc_one_shot.compute_imputed(n_epochs=n_epochs_one_shot)
brain_large_one_shot = Benchmarkable(
trainer_fname="trainer_brain_large_one_shot",
model_fname="model_brain_large_one_shot",
name="Brain Large default",
is_one_shot=True,
)
brain_large_one_shot.compute_imputed(n_epochs=n_epochs_brain_large)
training: 100%|██████████| 400/400 [02:32<00:00, 2.63it/s]
Median of Median: 2.3032
Mean of Median for each cell: 3.2574
training: 100%|██████████| 400/400 [15:54<00:00, 2.38s/it]
Median of Median: 0.8406
Mean of Median for each cell: 0.9256
training: 100%|██████████| 50/50 [2:27:55<00:00, 177.71s/it]
Median of Median: 0.0000
Mean of Median for each cell: 0.4581
10.11. Hyperparameter space DataFrame¶
Our helper class allows us to get a dataframe per experiment resuming the results of each trial.
[6]:
cortex_df = cortex.get_param_df()
cortex_df.to_csv("cortex_df")
cortex_df
[6]:
| marginal_ll | n_layers | n_hidden | n_latent | reconstruction_loss | dropout_rate | lr | n_epochs | n_params | run index | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1218.52 | 1 | 256 | 10 | zinb | 0.1 | 0.01 | 248 | 290816 | 92 |
| 2 | 1218.7 | 1 | 128 | 12 | zinb | 0.1 | 0.01 | 382 | 145920 | 80 |
| 3 | 1219.7 | 1 | 256 | 10 | zinb | 0.1 | 0.01 | 365 | 290816 | 85 |
| 4 | 1220.06 | 1 | 256 | 10 | zinb | 0.1 | 0.01 | 275 | 290816 | 91 |
| 5 | 1223.09 | 1 | 128 | 10 | zinb | 0.1 | 0.01 | 440 | 145408 | 83 |
| 6 | 1223.2 | 1 | 128 | 12 | zinb | 0.5 | 0.005 | 703 | 145920 | 38 |
| 7 | 1223.53 | 1 | 256 | 10 | zinb | 0.1 | 0.001 | 514 | 290816 | 97 |
| 8 | 1223.94 | 1 | 128 | 12 | zinb | 0.5 | 0.01 | 542 | 145920 | 74 |
| 9 | 1224.37 | 1 | 128 | 12 | zinb | 0.5 | 0.01 | 524 | 145920 | 76 |
| 10 | 1224.37 | 1 | 128 | 12 | zinb | 0.5 | 0.01 | 497 | 145920 | 71 |
| 11 | 1225.6 | 1 | 128 | 6 | zinb | 0.5 | 0.005 | 596 | 144384 | 24 |
| 12 | 1225.73 | 1 | 128 | 6 | zinb | 0.5 | 0.01 | 565 | 144384 | 25 |
| 13 | 1225.76 | 1 | 128 | 14 | zinb | 0.5 | 0.01 | 421 | 146432 | 31 |
| 14 | 1225.83 | 1 | 128 | 13 | zinb | 0.5 | 0.01 | 560 | 146176 | 28 |
| 15 | 1225.86 | 1 | 128 | 6 | zinb | 0.5 | 0.01 | 496 | 144384 | 67 |
| 16 | 1225.9 | 1 | 128 | 6 | zinb | 0.5 | 0.01 | 512 | 144384 | 66 |
| 17 | 1226.23 | 1 | 128 | 6 | zinb | 0.5 | 0.01 | 508 | 144384 | 68 |
| 18 | 1226.23 | 1 | 128 | 6 | zinb | 0.1 | 0.01 | 388 | 144384 | 23 |
| 19 | 1226.52 | 1 | 128 | 10 | zinb | 0.5 | 0.01 | 491 | 145408 | 29 |
| 20 | 1226.63 | 1 | 128 | 6 | zinb | 0.5 | 0.01 | 554 | 144384 | 70 |
| 21 | 1226.65 | 1 | 128 | 6 | zinb | 0.5 | 0.01 | 458 | 144384 | 27 |
| 22 | 1226.71 | 1 | 128 | 6 | zinb | 0.5 | 0.01 | 536 | 144384 | 69 |
| 23 | 1226.93 | 1 | 128 | 14 | zinb | 0.5 | 0.005 | 596 | 146432 | 59 |
| 24 | 1227.18 | 1 | 128 | 6 | zinb | 0.5 | 0.01 | 493 | 144384 | 26 |
| 25 | 1227.33 | 1 | 128 | 6 | zinb | 0.5 | 0.005 | 702 | 144384 | 22 |
| 26 | 1227.9 | 2 | 256 | 10 | zinb | 0.1 | 0.01 | 266 | 421888 | 89 |
| 27 | 1228.21 | 1 | 128 | 6 | zinb | 0.1 | 0.005 | 457 | 144384 | 21 |
| 28 | 1228.99 | 1 | 256 | 6 | zinb | 0.1 | 0.005 | 295 | 288768 | 17 |
| 29 | 1230.29 | 1 | 256 | 7 | zinb | 0.5 | 0.005 | 530 | 289280 | 47 |
| 30 | 1230.48 | 1 | 256 | 10 | nb | 0.1 | 0.01 | 335 | 290816 | 88 |
| 31 | 1230.94 | 2 | 256 | 14 | zinb | 0.1 | 0.01 | 405 | 423936 | 96 |
| 32 | 1231.77 | 2 | 128 | 11 | zinb | 0.3 | 0.005 | 580 | 178432 | 16 |
| 33 | 1238.09 | 1 | 256 | 8 | nb | 0.5 | 0.005 | 877 | 289792 | 50 |
| 34 | 1238.13 | 1 | 128 | 12 | nb | 0.1 | 0.001 | 643 | 145920 | 84 |
| 35 | 1238.17 | 4 | 256 | 8 | zinb | 0.1 | 0.01 | 289 | 683008 | 95 |
| 36 | 1238.57 | 4 | 256 | 10 | zinb | 0.1 | 0.01 | 343 | 684032 | 87 |
| 37 | 1238.77 | 4 | 256 | 5 | zinb | 0.1 | 0.001 | 499 | 681472 | 9 |
| 38 | 1238.96 | 1 | 128 | 6 | zinb | 0.7 | 0.005 | 664 | 144384 | 62 |
| 39 | 1239.09 | 1 | 128 | 6 | zinb | 0.5 | 0.001 | 993 | 144384 | 30 |
| 40 | 1240.54 | 1 | 64 | 15 | zinb | 0.5 | 0.01 | 527 | 73344 | 39 |
| 41 | 1241.08 | 3 | 256 | 10 | zinb | 0.1 | 0.01 | 357 | 552960 | 90 |
| 42 | 1241.39 | 3 | 128 | 5 | zinb | 0.1 | 0.001 | 661 | 209664 | 64 |
| 43 | 1241.46 | 4 | 256 | 11 | zinb | 0.1 | 0.0005 | 550 | 684544 | 19 |
| 44 | 1241.7 | 1 | 64 | 12 | zinb | 0.5 | 0.01 | 500 | 72960 | 77 |
| 45 | 1241.74 | 1 | 256 | 5 | nb | 0.1 | 0.005 | 464 | 288256 | 53 |
| 46 | 1242.13 | 2 | 128 | 12 | zinb | 0.3 | 0.01 | 484 | 178688 | 81 |
| 47 | 1242.96 | 3 | 64 | 13 | zinb | 0.1 | 0.01 | 546 | 89472 | 82 |
| 48 | 1244.33 | 2 | 256 | 9 | nb | 0.5 | 0.005 | 803 | 421376 | 63 |
| 49 | 1245.95 | 2 | 256 | 7 | nb | 0.1 | 0.001 | 458 | 420352 | 58 |
| 50 | 1253.61 | 3 | 256 | 9 | nb | 0.1 | 0.0005 | 540 | 552448 | 98 |
| 51 | 1254.13 | 5 | 256 | 7 | zinb | 0.1 | 0.01 | 407 | 813568 | 94 |
| 52 | 1257.09 | 3 | 256 | 12 | nb | 0.5 | 0.005 | 685 | 553984 | 43 |
| 53 | 1260.27 | 3 | 256 | 7 | zinb | 0.3 | 0.0005 | 669 | 551424 | 8 |
| 54 | 1260.59 | 3 | 256 | 14 | zinb | 0.3 | 0.0005 | 619 | 555008 | 18 |
| 55 | 1261.66 | 4 | 128 | 12 | zinb | 0.3 | 0.005 | 799 | 244224 | 41 |
| 56 | 1262.43 | 3 | 128 | 10 | zinb | 0.5 | 0.005 | 606 | 210944 | 57 |
| 57 | 1263.45 | 5 | 128 | 10 | zinb | 0.1 | 0.0005 | 674 | 276480 | 86 |
| 58 | 1263.61 | 1 | 64 | 11 | zinb | 0.7 | 0.005 | 887 | 72832 | 55 |
| 59 | 1265.92 | 2 | 128 | 9 | zinb | 0.3 | 0.0005 | 783 | 177920 | 51 |
| 60 | 1269.22 | 4 | 128 | 12 | zinb | 0.5 | 0.005 | 606 | 244224 | 73 |
| 61 | 1270.39 | 5 | 128 | 12 | zinb | 0.5 | 0.01 | 599 | 276992 | 78 |
| 62 | 1270.91 | 4 | 128 | 12 | zinb | 0.5 | 0.01 | 506 | 244224 | 32 |
| 63 | 1274.21 | 4 | 128 | 13 | zinb | 0.3 | 0.0005 | 935 | 244480 | 56 |
| 64 | 1274.98 | 1 | 64 | 7 | nb | 0.7 | 0.005 | 640 | 72320 | 6 |
| 65 | 1282.63 | 4 | 128 | 14 | nb | 0.5 | 0.01 | 583 | 244736 | 0 |
| 66 | 1283.68 | 5 | 64 | 15 | nb | 0.1 | 0.0005 | 735 | 106112 | 3 |
| 67 | 1286.75 | 4 | 256 | 12 | nb | 0.7 | 0.01 | 590 | 685056 | 79 |
| 68 | 1286.77 | 1 | 128 | 13 | zinb | 0.9 | 0.005 | 495 | 146176 | 44 |
| 69 | 1287.13 | 1 | 128 | 12 | zinb | 0.9 | 0.005 | 566 | 145920 | 75 |
| 70 | 1287.57 | 3 | 128 | 5 | nb | 0.3 | 0.001 | 540 | 209664 | 46 |
| 71 | 1291.13 | 5 | 128 | 15 | nb | 0.5 | 0.005 | 986 | 277760 | 34 |
| 72 | 1299.72 | 5 | 128 | 11 | zinb | 0.3 | 0.0005 | 768 | 276736 | 65 |
| 73 | 1306.11 | 1 | 128 | 6 | zinb | 0.5 | 0.001 | 994 | 144384 | 35 |
| 74 | 1319.24 | 2 | 256 | 12 | zinb | 0.9 | 0.005 | 637 | 422912 | 7 |
| 75 | 1321.87 | 5 | 128 | 11 | zinb | 0.1 | 0.0001 | 998 | 276736 | 48 |
| 76 | 1335.01 | 5 | 64 | 12 | zinb | 0.7 | 0.005 | 382 | 105728 | 42 |
| 77 | 1345.81 | 5 | 64 | 5 | nb | 0.3 | 0.0005 | 741 | 104832 | 10 |
| 78 | 1349.62 | 2 | 128 | 12 | nb | 0.9 | 0.005 | 705 | 178688 | 40 |
| 79 | 1370.89 | 2 | 64 | 8 | nb | 0.9 | 0.005 | 526 | 80640 | 12 |
| 80 | 1373.79 | 1 | 128 | 9 | zinb | 0.5 | 0.0001 | 949 | 145152 | 33 |
| 81 | 1391.54 | 2 | 64 | 15 | nb | 0.3 | 0.0001 | 999 | 81536 | 4 |
| 82 | 1398.38 | 1 | 128 | 15 | zinb | 0.5 | 0.0001 | 769 | 146688 | 72 |
| 83 | 1399.38 | 1 | 256 | 5 | zinb | 0.1 | 0.0001 | 987 | 288256 | 99 |
| 84 | 1419.98 | 1 | 256 | 10 | nb | 0.1 | 0.0001 | 903 | 290816 | 93 |
| 85 | 1436.06 | 5 | 128 | 15 | zinb | 0.5 | 0.0001 | 774 | 277760 | 54 |
| 86 | 1463.22 | 4 | 128 | 8 | zinb | 0.9 | 0.01 | 382 | 243200 | 36 |
| 87 | 1510.45 | 3 | 128 | 13 | zinb | 0.7 | 0.0005 | 168 | 211712 | 14 |
| 88 | 1512.93 | 3 | 128 | 8 | zinb | 0.7 | 0.0005 | 151 | 210432 | 5 |
| 89 | 1523.67 | 5 | 128 | 5 | zinb | 0.7 | 0.0005 | 257 | 275200 | 11 |
| 90 | 1542.96 | 4 | 256 | 9 | zinb | 0.7 | 0.0001 | 482 | 683520 | 2 |
| 91 | 1554.98 | 2 | 64 | 10 | zinb | 0.7 | 0.005 | 256 | 80896 | 45 |
| 92 | 1559.01 | 5 | 64 | 7 | nb | 0.5 | 0.0001 | 457 | 105088 | 37 |
| 93 | 1601.53 | 3 | 64 | 10 | nb | 0.7 | 0.001 | 88 | 89088 | 15 |
| 94 | 1612.9 | 4 | 64 | 14 | zinb | 0.7 | 0.005 | 71 | 97792 | 49 |
| 95 | 1615.22 | 2 | 256 | 9 | nb | 0.9 | 0.0001 | 197 | 421376 | 20 |
| 96 | 1746.25 | 3 | 128 | 12 | zinb | 0.9 | 0.001 | 134 | 211456 | 52 |
| 97 | 1818.82 | 1 | 64 | 12 | zinb | 0.9 | 0.0005 | 54 | 72960 | 60 |
| 98 | 6574.57 | 1 | 128 | 8 | zinb | 0.5 | 0.0001 | 4 | 144896 | 61 |
| 99 | 10680.4 | 5 | 64 | 12 | zinb | 0.3 | 0.0001 | 2 | 105728 | 1 |
| 100 | NaN | 2 | 64 | 6 | zinb | 0.9 | 0.0001 | 31 | 80384 | 13 |
[7]:
pbmc_df = pbmc.get_param_df()
pbmc_df.to_csv("pbmc_df")
pbmc_df
[7]:
| marginal_ll | n_layers | n_hidden | n_latent | reconstruction_loss | dropout_rate | lr | n_epochs | n_params | run index | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1323.79 | 1 | 128 | 10 | nb | 0.3 | 0.01 | 160 | 859136 | 29 |
| 2 | 1323.88 | 1 | 128 | 13 | nb | 0.3 | 0.005 | 238 | 859904 | 84 |
| 3 | 1324.08 | 1 | 128 | 15 | nb | 0.3 | 0.01 | 172 | 860416 | 37 |
| 4 | 1324.1 | 1 | 128 | 14 | nb | 0.3 | 0.005 | 275 | 860160 | 68 |
| 5 | 1324.24 | 1 | 128 | 14 | nb | 0.3 | 0.005 | 271 | 860160 | 65 |
| 6 | 1324.4 | 1 | 128 | 14 | nb | 0.3 | 0.005 | 196 | 860160 | 61 |
| 7 | 1324.53 | 1 | 128 | 13 | zinb | 0.3 | 0.001 | 411 | 859904 | 90 |
| 8 | 1324.55 | 1 | 128 | 6 | zinb | 0.3 | 0.001 | 419 | 858112 | 75 |
| 9 | 1324.58 | 1 | 256 | 8 | nb | 0.3 | 0.01 | 141 | 1717248 | 92 |
| 10 | 1324.62 | 1 | 128 | 11 | nb | 0.3 | 0.005 | 227 | 859392 | 70 |
| 11 | 1324.68 | 1 | 128 | 5 | nb | 0.1 | 0.01 | 180 | 857856 | 97 |
| 12 | 1324.74 | 1 | 128 | 13 | nb | 0.3 | 0.0005 | 624 | 859904 | 88 |
| 13 | 1324.77 | 1 | 128 | 14 | nb | 0.3 | 0.005 | 241 | 860160 | 67 |
| 14 | 1324.79 | 1 | 128 | 10 | nb | 0.1 | 0.001 | 313 | 859136 | 82 |
| 15 | 1324.81 | 1 | 128 | 14 | nb | 0.3 | 0.005 | 231 | 860160 | 66 |
| 16 | 1324.82 | 1 | 128 | 14 | nb | 0.3 | 0.005 | 230 | 860160 | 69 |
| 17 | 1324.83 | 2 | 128 | 9 | nb | 0.3 | 0.01 | 162 | 891648 | 22 |
| 18 | 1324.89 | 1 | 128 | 10 | zinb | 0.1 | 0.01 | 169 | 859136 | 59 |
| 19 | 1324.91 | 1 | 128 | 9 | zinb | 0.3 | 0.01 | 175 | 858880 | 28 |
| 20 | 1325.03 | 1 | 128 | 13 | nb | 0.5 | 0.001 | 468 | 859904 | 54 |
| 21 | 1325.08 | 1 | 128 | 10 | nb | 0.3 | 0.005 | 273 | 859136 | 71 |
| 22 | 1325.16 | 1 | 128 | 9 | zinb | 0.3 | 0.01 | 201 | 858880 | 30 |
| 23 | 1325.17 | 1 | 128 | 14 | nb | 0.3 | 0.01 | 200 | 860160 | 77 |
| 24 | 1325.28 | 1 | 128 | 13 | nb | 0.3 | 0.01 | 204 | 859904 | 72 |
| 25 | 1325.53 | 2 | 128 | 5 | zinb | 0.3 | 0.01 | 138 | 890624 | 23 |
| 26 | 1325.54 | 1 | 64 | 14 | nb | 0.3 | 0.01 | 225 | 430080 | 50 |
| 27 | 1325.55 | 2 | 128 | 15 | nb | 0.3 | 0.005 | 173 | 893184 | 73 |
| 28 | 1325.57 | 3 | 128 | 11 | nb | 0.1 | 0.01 | 165 | 924928 | 52 |
| 29 | 1325.62 | 2 | 128 | 5 | zinb | 0.3 | 0.01 | 175 | 890624 | 27 |
| 30 | 1325.68 | 1 | 256 | 15 | nb | 0.3 | 0.001 | 287 | 1720832 | 98 |
| 31 | 1325.83 | 2 | 128 | 9 | nb | 0.3 | 0.01 | 151 | 891648 | 26 |
| 32 | 1326.03 | 2 | 128 | 9 | zinb | 0.3 | 0.01 | 168 | 891648 | 25 |
| 33 | 1326.03 | 2 | 128 | 14 | zinb | 0.5 | 0.001 | 376 | 892928 | 15 |
| 34 | 1326.04 | 1 | 128 | 15 | nb | 0.5 | 0.0005 | 596 | 860416 | 48 |
| 35 | 1326.07 | 2 | 128 | 5 | nb | 0.3 | 0.01 | 192 | 890624 | 20 |
| 36 | 1326.12 | 1 | 64 | 10 | nb | 0.3 | 0.01 | 287 | 429568 | 31 |
| 37 | 1326.16 | 2 | 128 | 9 | nb | 0.5 | 0.001 | 460 | 891648 | 24 |
| 38 | 1326.18 | 2 | 128 | 7 | nb | 0.5 | 0.001 | 406 | 891136 | 21 |
| 39 | 1326.28 | 2 | 256 | 14 | nb | 0.1 | 0.01 | 109 | 1851392 | 19 |
| 40 | 1326.65 | 3 | 256 | 15 | nb | 0.3 | 0.005 | 189 | 1982976 | 87 |
| 41 | 1327.05 | 1 | 256 | 15 | nb | 0.3 | 0.0005 | 418 | 1720832 | 43 |
| 42 | 1327.06 | 1 | 64 | 7 | nb | 0.3 | 0.01 | 207 | 429184 | 35 |
| 43 | 1327.47 | 3 | 64 | 11 | nb | 0.1 | 0.005 | 281 | 446080 | 38 |
| 44 | 1327.52 | 4 | 128 | 5 | nb | 0.3 | 0.01 | 173 | 956160 | 12 |
| 45 | 1327.55 | 2 | 256 | 6 | zinb | 0.5 | 0.0005 | 484 | 1847296 | 9 |
| 46 | 1327.81 | 1 | 128 | 15 | nb | 0.7 | 0.001 | 454 | 860416 | 39 |
| 47 | 1328 | 5 | 128 | 6 | nb | 0.1 | 0.0005 | 395 | 989184 | 16 |
| 48 | 1328.15 | 1 | 128 | 14 | zinb | 0.7 | 0.01 | 191 | 860160 | 85 |
| 49 | 1328.17 | 1 | 128 | 7 | zinb | 0.7 | 0.01 | 339 | 858368 | 62 |
| 50 | 1328.25 | 5 | 128 | 13 | nb | 0.3 | 0.01 | 279 | 990976 | 45 |
| 51 | 1328.35 | 5 | 64 | 8 | zinb | 0.1 | 0.001 | 383 | 462080 | 7 |
| 52 | 1328.36 | 1 | 64 | 10 | nb | 0.5 | 0.005 | 292 | 429568 | 42 |
| 53 | 1328.52 | 5 | 128 | 6 | nb | 0.3 | 0.001 | 431 | 989184 | 58 |
| 54 | 1328.68 | 3 | 256 | 15 | zinb | 0.5 | 0.001 | 383 | 1982976 | 3 |
| 55 | 1328.78 | 5 | 128 | 10 | zinb | 0.3 | 0.005 | 245 | 990208 | 53 |
| 56 | 1328.82 | 3 | 64 | 8 | nb | 0.3 | 0.005 | 303 | 445696 | 11 |
| 57 | 1328.82 | 4 | 128 | 8 | zinb | 0.3 | 0.0005 | 593 | 956928 | 13 |
| 58 | 1328.99 | 4 | 256 | 10 | nb | 0.5 | 0.01 | 258 | 2111488 | 76 |
| 59 | 1329.02 | 3 | 128 | 6 | zinb | 0.3 | 0.0005 | 541 | 923648 | 95 |
| 60 | 1329.1 | 3 | 64 | 12 | nb | 0.3 | 0.0005 | 652 | 446208 | 57 |
| 61 | 1329.11 | 3 | 64 | 10 | nb | 0.1 | 0.001 | 409 | 445952 | 46 |
| 62 | 1329.17 | 4 | 256 | 6 | nb | 0.3 | 0.0005 | 431 | 2109440 | 51 |
| 63 | 1329.36 | 1 | 128 | 12 | nb | 0.7 | 0.01 | 286 | 859648 | 44 |
| 64 | 1330.12 | 1 | 128 | 12 | nb | 0.1 | 0.0001 | 923 | 859648 | 74 |
| 65 | 1330.57 | 4 | 128 | 13 | nb | 0.5 | 0.01 | 246 | 958208 | 91 |
| 66 | 1330.59 | 4 | 128 | 7 | nb | 0.5 | 0.005 | 268 | 956672 | 99 |
| 67 | 1331.04 | 2 | 128 | 12 | nb | 0.7 | 0.005 | 452 | 892416 | 93 |
| 68 | 1331.31 | 5 | 256 | 9 | nb | 0.3 | 0.0005 | 400 | 2242048 | 81 |
| 69 | 1331.92 | 1 | 128 | 15 | nb | 0.3 | 0.0001 | 999 | 860416 | 41 |
| 70 | 1332.08 | 1 | 128 | 11 | nb | 0.3 | 0.0001 | 991 | 859392 | 32 |
| 71 | 1333.71 | 5 | 64 | 8 | nb | 0.3 | 0.01 | 321 | 462080 | 63 |
| 72 | 1334.2 | 3 | 128 | 15 | zinb | 0.3 | 0.0001 | 998 | 925952 | 80 |
| 73 | 1334.2 | 3 | 256 | 15 | zinb | 0.5 | 0.0001 | 987 | 1982976 | 4 |
| 74 | 1335.42 | 1 | 128 | 10 | nb | 0.1 | 0.0001 | 822 | 859136 | 34 |
| 75 | 1335.43 | 4 | 256 | 15 | nb | 0.7 | 0.01 | 352 | 2114048 | 55 |
| 76 | 1335.46 | 4 | 128 | 11 | nb | 0.1 | 0.0001 | 992 | 957696 | 1 |
| 77 | 1336.01 | 1 | 256 | 13 | nb | 0.7 | 0.0001 | 996 | 1719808 | 2 |
| 78 | 1336.85 | 4 | 256 | 15 | nb | 0.5 | 0.0001 | 995 | 2114048 | 60 |
| 79 | 1337.03 | 4 | 256 | 13 | zinb | 0.7 | 0.0005 | 585 | 2113024 | 18 |
| 80 | 1337.34 | 2 | 64 | 5 | nb | 0.7 | 0.005 | 336 | 437120 | 78 |
| 81 | 1337.93 | 1 | 128 | 14 | zinb | 0.9 | 0.001 | 500 | 860160 | 0 |
| 82 | 1338.55 | 5 | 64 | 13 | nb | 0.1 | 0.0001 | 997 | 462720 | 89 |
| 83 | 1338.56 | 1 | 256 | 15 | nb | 0.9 | 0.01 | 262 | 1720832 | 40 |
| 84 | 1339.85 | 4 | 64 | 11 | nb | 0.5 | 0.01 | 374 | 454272 | 83 |
| 85 | 1341.08 | 5 | 128 | 9 | nb | 0.3 | 0.0001 | 991 | 989952 | 96 |
| 86 | 1347.57 | 1 | 128 | 8 | zinb | 0.9 | 0.01 | 51 | 858624 | 56 |
| 87 | 1348.94 | 1 | 128 | 7 | nb | 0.9 | 0.01 | 57 | 858368 | 79 |
| 88 | 1350.36 | 1 | 128 | 10 | nb | 0.9 | 0.01 | 54 | 859136 | 33 |
| 89 | 1352.03 | 4 | 256 | 12 | zinb | 0.9 | 0.005 | 344 | 2112512 | 6 |
| 90 | 1353.97 | 5 | 64 | 5 | nb | 0.7 | 0.01 | 390 | 461696 | 14 |
| 91 | 1359.17 | 5 | 64 | 13 | nb | 0.7 | 0.0005 | 608 | 462720 | 8 |
| 92 | 1360.53 | 4 | 256 | 8 | nb | 0.9 | 0.005 | 129 | 2110464 | 47 |
| 93 | 1362.3 | 4 | 256 | 6 | zinb | 0.9 | 0.005 | 115 | 2109440 | 17 |
| 94 | 1362.45 | 1 | 64 | 10 | nb | 0.9 | 0.01 | 67 | 429568 | 94 |
| 95 | 1363.52 | 3 | 128 | 5 | nb | 0.9 | 0.0005 | 516 | 923392 | 64 |
| 96 | 1365.34 | 5 | 128 | 14 | nb | 0.7 | 0.005 | 69 | 991232 | 10 |
| 97 | 1365.92 | 3 | 256 | 10 | zinb | 0.9 | 0.0001 | 999 | 1980416 | 5 |
| 98 | 1368.19 | 2 | 128 | 13 | nb | 0.9 | 0.01 | 51 | 892672 | 86 |
| 99 | 1509.34 | 5 | 128 | 7 | nb | 0.7 | 0.0001 | 40 | 989440 | 49 |
| 100 | 1595.89 | 3 | 128 | 12 | nb | 0.9 | 0.0001 | 130 | 925184 | 36 |
[9]:
brain_large_df = brain_large.get_param_df()
brain_large_df.to_csv("brain_large_df")
brain_large_df
[9]:
| marginal_ll | n_layers | n_hidden | n_latent | reconstruction_loss | dropout_rate | lr | n_epochs | n_params | run index | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 138.77 | 1 | 256 | 8 | zinb | 0.1 | 0.001 | 50 | 372736 | 67 |
| 2 | 138.779 | 1 | 256 | 15 | zinb | 0.1 | 0.001 | 46 | 376320 | 24 |
| 3 | 138.794 | 1 | 256 | 11 | zinb | 0.1 | 0.001 | 48 | 374272 | 73 |
| 4 | 138.798 | 1 | 256 | 8 | zinb | 0.1 | 0.001 | 45 | 372736 | 38 |
| 5 | 138.81 | 1 | 256 | 10 | zinb | 0.1 | 0.001 | 49 | 373760 | 70 |
| 6 | 138.828 | 1 | 256 | 8 | zinb | 0.1 | 0.001 | 46 | 372736 | 66 |
| 7 | 138.852 | 1 | 256 | 8 | zinb | 0.1 | 0.001 | 48 | 372736 | 41 |
| 8 | 138.894 | 1 | 256 | 8 | zinb | 0.1 | 0.001 | 42 | 372736 | 52 |
| 9 | 138.899 | 1 | 256 | 8 | zinb | 0.1 | 0.001 | 47 | 372736 | 26 |
| 10 | 138.902 | 1 | 256 | 12 | zinb | 0.1 | 0.005 | 47 | 374784 | 62 |
| 11 | 138.904 | 1 | 256 | 12 | zinb | 0.1 | 0.0001 | 47 | 374784 | 81 |
| 12 | 138.91 | 1 | 256 | 8 | zinb | 0.1 | 0.001 | 49 | 372736 | 47 |
| 13 | 138.911 | 1 | 256 | 9 | zinb | 0.1 | 0.0001 | 45 | 373248 | 74 |
| 14 | 138.914 | 1 | 256 | 8 | zinb | 0.1 | 0.001 | 46 | 372736 | 68 |
| 15 | 138.971 | 1 | 256 | 8 | zinb | 0.1 | 0.001 | 46 | 372736 | 65 |
| 16 | 139.126 | 1 | 128 | 13 | nb | 0.1 | 0.0005 | 49 | 187648 | 33 |
| 17 | 139.129 | 1 | 128 | 8 | nb | 0.1 | 0.001 | 48 | 186368 | 64 |
| 18 | 139.13 | 2 | 256 | 13 | zinb | 0.1 | 0.0005 | 43 | 506368 | 76 |
| 19 | 139.141 | 1 | 256 | 11 | zinb | 0.1 | 0.005 | 46 | 374272 | 93 |
| 20 | 139.143 | 1 | 128 | 13 | nb | 0.1 | 0.0005 | 49 | 187648 | 16 |
| 21 | 139.163 | 1 | 256 | 6 | zinb | 0.1 | 0.001 | 47 | 371712 | 69 |
| 22 | 139.19 | 1 | 256 | 14 | nb | 0.3 | 0.0005 | 46 | 375808 | 35 |
| 23 | 139.227 | 2 | 256 | 15 | zinb | 0.1 | 0.001 | 42 | 507392 | 99 |
| 24 | 139.251 | 1 | 128 | 11 | zinb | 0.1 | 0.001 | 44 | 187136 | 90 |
| 25 | 139.267 | 1 | 256 | 14 | zinb | 0.3 | 0.001 | 46 | 375808 | 72 |
| 26 | 139.304 | 1 | 256 | 14 | zinb | 0.3 | 0.001 | 46 | 375808 | 21 |
| 27 | 139.333 | 1 | 256 | 9 | zinb | 0.3 | 0.001 | 48 | 373248 | 88 |
| 28 | 139.344 | 1 | 256 | 8 | nb | 0.3 | 0.001 | 49 | 372736 | 58 |
| 29 | 139.422 | 1 | 64 | 9 | zinb | 0.1 | 0.001 | 48 | 93312 | 45 |
| 30 | 139.454 | 2 | 256 | 8 | nb | 0.1 | 0.001 | 45 | 503808 | 82 |
| 31 | 139.508 | 2 | 128 | 10 | zinb | 0.1 | 0.001 | 48 | 219648 | 25 |
| 32 | 139.528 | 2 | 256 | 8 | zinb | 0.1 | 0.0001 | 47 | 503808 | 89 |
| 33 | 139.549 | 1 | 64 | 11 | nb | 0.1 | 0.0001 | 48 | 93568 | 97 |
| 34 | 139.59 | 1 | 64 | 13 | nb | 0.1 | 0.0005 | 48 | 93824 | 37 |
| 35 | 139.599 | 2 | 128 | 7 | nb | 0.1 | 0.0005 | 49 | 218880 | 34 |
| 36 | 139.742 | 3 | 256 | 11 | zinb | 0.1 | 0.001 | 49 | 636416 | 91 |
| 37 | 139.749 | 3 | 256 | 8 | zinb | 0.1 | 0.005 | 48 | 634880 | 39 |
| 38 | 139.803 | 3 | 256 | 14 | zinb | 0.1 | 0.0005 | 48 | 637952 | 83 |
| 39 | 139.825 | 3 | 256 | 7 | zinb | 0.1 | 0.001 | 44 | 634368 | 98 |
| 40 | 139.906 | 1 | 256 | 7 | zinb | 0.5 | 0.001 | 48 | 372224 | 27 |
| 41 | 139.953 | 2 | 128 | 15 | zinb | 0.1 | 0.001 | 46 | 220928 | 15 |
| 42 | 139.974 | 1 | 256 | 5 | zinb | 0.5 | 0.001 | 43 | 371200 | 3 |
| 43 | 139.975 | 1 | 256 | 10 | zinb | 0.5 | 0.005 | 48 | 373760 | 85 |
| 44 | 139.996 | 1 | 256 | 5 | zinb | 0.5 | 0.001 | 49 | 371200 | 20 |
| 45 | 140.024 | 1 | 64 | 13 | zinb | 0.1 | 0.01 | 45 | 93824 | 86 |
| 46 | 140.034 | 2 | 256 | 6 | zinb | 0.3 | 0.0005 | 44 | 502784 | 23 |
| 47 | 140.093 | 1 | 256 | 14 | nb | 0.3 | 0.01 | 47 | 375808 | 36 |
| 48 | 140.155 | 2 | 256 | 14 | zinb | 0.1 | 0.01 | 38 | 506880 | 31 |
| 49 | 140.227 | 1 | 64 | 15 | zinb | 0.3 | 0.001 | 48 | 94080 | 80 |
| 50 | 140.238 | 2 | 128 | 6 | zinb | 0.3 | 0.0005 | 42 | 218624 | 0 |
| 51 | 140.389 | 1 | 256 | 11 | zinb | 0.5 | 0.01 | 47 | 374272 | 94 |
| 52 | 140.392 | 4 | 256 | 15 | zinb | 0.1 | 0.001 | 46 | 769536 | 57 |
| 53 | 140.466 | 3 | 256 | 10 | zinb | 0.1 | 0.01 | 49 | 635904 | 43 |
| 54 | 140.558 | 3 | 64 | 11 | zinb | 0.1 | 0.005 | 49 | 109952 | 48 |
| 55 | 140.596 | 1 | 256 | 5 | zinb | 0.7 | 0.001 | 48 | 371200 | 60 |
| 56 | 140.603 | 3 | 256 | 15 | zinb | 0.3 | 0.0001 | 47 | 638464 | 53 |
| 57 | 140.61 | 4 | 128 | 15 | zinb | 0.1 | 0.001 | 47 | 286464 | 29 |
| 58 | 140.612 | 1 | 128 | 15 | zinb | 0.5 | 0.001 | 42 | 188160 | 32 |
| 59 | 140.623 | 4 | 64 | 12 | zinb | 0.1 | 0.001 | 47 | 118272 | 51 |
| 60 | 140.661 | 1 | 256 | 15 | zinb | 0.7 | 0.005 | 39 | 376320 | 77 |
| 61 | 140.669 | 1 | 256 | 6 | zinb | 0.7 | 0.001 | 38 | 371712 | 54 |
| 62 | 140.734 | 3 | 64 | 5 | zinb | 0.1 | 0.005 | 49 | 109184 | 9 |
| 63 | 140.753 | 1 | 256 | 15 | zinb | 0.7 | 0.0001 | 48 | 376320 | 49 |
| 64 | 140.829 | 2 | 256 | 8 | zinb | 0.5 | 0.0005 | 49 | 503808 | 71 |
| 65 | 140.856 | 2 | 256 | 5 | zinb | 0.5 | 0.0005 | 47 | 502272 | 22 |
| 66 | 140.958 | 2 | 128 | 15 | nb | 0.3 | 0.001 | 49 | 220928 | 11 |
| 67 | 141.075 | 2 | 128 | 5 | zinb | 0.5 | 0.001 | 48 | 218368 | 28 |
| 68 | 141.513 | 5 | 128 | 5 | zinb | 0.1 | 0.001 | 49 | 316672 | 75 |
| 69 | 141.649 | 5 | 256 | 12 | nb | 0.1 | 0.005 | 49 | 899072 | 12 |
| 70 | 141.751 | 4 | 128 | 5 | zinb | 0.3 | 0.001 | 40 | 283904 | 96 |
| 71 | 141.792 | 5 | 64 | 10 | zinb | 0.1 | 0.005 | 39 | 126208 | 55 |
| 72 | 141.858 | 1 | 128 | 13 | zinb | 0.7 | 0.0001 | 48 | 187648 | 10 |
| 73 | 141.888 | 4 | 256 | 9 | nb | 0.5 | 0.0005 | 49 | 766464 | 8 |
| 74 | 141.906 | 3 | 64 | 9 | zinb | 0.1 | 0.0001 | 47 | 109696 | 59 |
| 75 | 141.927 | 5 | 256 | 15 | nb | 0.5 | 0.001 | 49 | 900608 | 61 |
| 76 | 141.986 | 3 | 128 | 8 | nb | 0.3 | 0.0005 | 49 | 251904 | 6 |
| 77 | 142.044 | 5 | 128 | 9 | zinb | 0.5 | 0.005 | 41 | 317696 | 30 |
| 78 | 142.138 | 3 | 64 | 6 | nb | 0.3 | 0.0005 | 49 | 109312 | 5 |
| 79 | 142.145 | 4 | 256 | 8 | zinb | 0.7 | 0.001 | 49 | 765952 | 40 |
| 80 | 142.154 | 3 | 256 | 8 | nb | 0.5 | 0.01 | 49 | 634880 | 78 |
| 81 | 142.165 | 1 | 256 | 15 | zinb | 0.9 | 0.001 | 44 | 376320 | 95 |
| 82 | 142.172 | 3 | 128 | 6 | nb | 0.5 | 0.005 | 45 | 251392 | 1 |
| 83 | 142.221 | 2 | 128 | 10 | zinb | 0.5 | 0.01 | 30 | 219648 | 7 |
| 84 | 142.365 | 5 | 256 | 15 | zinb | 0.7 | 0.0001 | 49 | 900608 | 44 |
| 85 | 142.373 | 5 | 256 | 13 | nb | 0.7 | 0.0001 | 49 | 899584 | 19 |
| 86 | 142.639 | 5 | 128 | 6 | zinb | 0.7 | 0.001 | 46 | 316928 | 84 |
| 87 | 143.32 | 1 | 64 | 7 | nb | 0.7 | 0.01 | 7 | 93056 | 2 |
| 88 | 143.498 | 1 | 256 | 6 | zinb | 0.9 | 0.005 | 3 | 371712 | 42 |
| 89 | 144.824 | 1 | 256 | 11 | zinb | 0.9 | 0.01 | 0 | 374272 | 56 |
| 90 | 146.517 | 5 | 64 | 13 | nb | 0.7 | 0.01 | 17 | 126592 | 14 |
| 91 | 146.626 | 5 | 64 | 11 | nb | 0.7 | 0.0005 | 44 | 126336 | 92 |
| 92 | 146.757 | 4 | 256 | 12 | zinb | 0.9 | 0.001 | 45 | 768000 | 46 |
| 93 | 146.837 | 4 | 128 | 7 | zinb | 0.9 | 0.001 | 33 | 284416 | 79 |
| 94 | 146.863 | 4 | 256 | 11 | nb | 0.9 | 0.001 | 42 | 767488 | 87 |
| 95 | 147.011 | 5 | 256 | 7 | zinb | 0.9 | 0.01 | 6 | 896512 | 50 |
| 96 | 147.021 | 2 | 128 | 12 | zinb | 0.9 | 0.001 | 47 | 220160 | 13 |
| 97 | 147.024 | 4 | 64 | 15 | zinb | 0.9 | 0.001 | 37 | 118656 | 4 |
| 98 | 147.369 | 4 | 64 | 11 | nb | 0.9 | 0.0001 | 43 | 118144 | 18 |
| 99 | 147.459 | 3 | 64 | 11 | nb | 0.9 | 0.0001 | 45 | 109952 | 17 |
| 100 | 148.457 | 4 | 64 | 7 | zinb | 0.9 | 0.01 | 0 | 117632 | 63 |
10.12. Best run DataFrame¶
Using the previous dataframes we are able to build one containing the best results along with the results obtained with the default parameters.
[18]:
cortex_best = cortex_df.iloc[0]
cortex_best.name = "Cortex tuned"
cortex_default = pd.Series(
[
cortex_one_shot.best_performance,
1, 128, 10, "zinb", 0.1, 0.001, 400, None, None
],
index=cortex_best.index
)
cortex_default.name = "Cortex default"
pbmc_best = pbmc_df.iloc[0]
pbmc_best.name = "Pbmc tuned"
pbmc_default = pd.Series(
[
pbmc_one_shot.best_performance,
1, 128, 10, "zinb", 0.1, 0.001, 400, None, None
],
index=pbmc_best.index
)
pbmc_default.name = "Pbmc default"
brain_large_best = brain_large_df.iloc[0]
brain_large_best.name = "Brain Large tuned"
brain_large_default = pd.Series(
[
brain_large_one_shot.best_performance,
1, 128, 10, "zinb", 0.1, 0.001, 400, None, None
],
index=brain_large_best.index
)
brain_large_default.name = "Brain Large default"
df_best = pd.concat(
[cortex_best,
cortex_default,
pbmc_best,
pbmc_default,
brain_large_best,
brain_large_default
],
axis=1
)
df_best = df_best.iloc[np.logical_not(np.isin(df_best.index, ["n_params", "run index"]))]
df_best
[18]:
| Cortex tuned | Cortex default | Pbmc tuned 16 GPUs | Pbmc default | Brain Large tuned | Brain Large default | |
|---|---|---|---|---|---|---|
| marginal_ll | 1218.52 | 1256.03 | 1323.44 | 1327.61 | 138.77 | 147.088 |
| n_layers | 1 | 1 | 1 | 1 | 1 | 1 |
| n_hidden | 256 | 128 | 256 | 128 | 256 | 128 |
| n_latent | 10 | 10 | 14 | 10 | 8 | 10 |
| reconstruction_loss | zinb | zinb | zinb | zinb | zinb | zinb |
| dropout_rate | 0.1 | 0.1 | 0.5 | 0.1 | 0.1 | 0.1 |
| lr | 0.01 | 0.001 | 0.01 | 0.001 | 0.001 | 0.001 |
| n_epochs | 248 | 400 | 170 | 400 | 50 | 400 |
10.13. Handy class to compare the results of each experiment¶
We use a second handy class to compare these results altogether. Specifically, the PlotBenchmarkable allows to retrieve: * A DataFrame containg the runtime information of each experiment. * A DataFrame comparint the different benchmarks (negative marginal LL, imputation) between tuned and default VAEs. * For each dataset, a plot aggregating the ELBO histories of each run.
[55]:
from notebooks.utils.autotune_advanced_notebook import PlotBenchmarkables
[56]:
tuned_benchmarkables = {
"cortex": cortex,
"pbmc": pbmc,
"brain large": brain_large,
}
one_shot_benchmarkables = {
"cortex": cortex_one_shot,
"pbmc": pbmc_one_shot,
"brain large": brain_large_one_shot
}
plotter = PlotBenchmarkables(
tuned_benchmarkables=tuned_benchmarkables,
one_shot_benchmarkables=one_shot_benchmarkables,
)
10.14. Runtime DataFrame¶
[27]:
df_runtime = plotter.get_runtime_dataframe()
df_runtime
[27]:
| Nb cells | Nb genes | Total GPU time | Total wall time | Number of trainings | Avg time per training | Avg epochs per training | Number of GPUs | Best epoch | Max epoch | |
|---|---|---|---|---|---|---|---|---|---|---|
| cortex | 3005 | 558 | 8:58:21.324546 | 9:02:20.162471 | 100 | 323.013 | 532.08 | 1 | 248 | 1000 |
| pbmc | 11990 | 3346 | 1 day, 23:24:59.874052 | 3:04:12.595907 | 100 | 1707 | 387.01 | 1 | 170 | 1000 |
| brain large | 1303182 | 720 | 12 days, 13:55:18.109345 | 21:38:48.882951 | 100 | 10869.2 | 43.51 | 16 | 50 | 50 |
10.15. Results DataFrame for best runs¶
[28]:
def highlight_min(data, color="yellow"):
attr = "background-color: {}".format(color)
if data.ndim == 1: # Series from .apply(axis=0) or axis=1
is_min = data == data.min()
return [attr if v else "" for v in is_min]
else: # from .apply(axis=None)
is_min = data == data.min().min()
return pd.DataFrame(np.where(is_min, attr, ""),
index=data.index, columns=data.columns)
[29]:
df_results = plotter.get_results_dataframe()
styler = df_results.style.apply(highlight_min, axis=0, subset=pd.IndexSlice["cortex", :])
styler = styler.apply(highlight_min, axis=0, subset=pd.IndexSlice["pbmc", :])
styler = styler.apply(highlight_min, axis=0, subset=pd.IndexSlice["brain large", :])
styler
[29]:
| Likelihood | Imputation score | |||||
|---|---|---|---|---|---|---|
| Held-out marginal ll | ELBO train | ELBO test | median | mean | ||
| cortex | tuned | 1218.52 | 1178.52 | 1231.16 | 2.08155 | 2.87502 |
| default | 1256.03 | 1224.18 | 1274.02 | 2.30317 | 3.25738 | |
| pbmc | tuned | 1323.44 | 1314.07 | 1328.04 | 0.83942 | 0.924637 |
| default | 1327.61 | 1309.9 | 1334.01 | 0.840617 | 0.925628 | |
| brain large | tuned | 138.77 | 141.783 | 141.899 | 0 | 0.392006 |
| default | 147.088 | 150.897 | 150.99 | 0 | 0.458067 | |
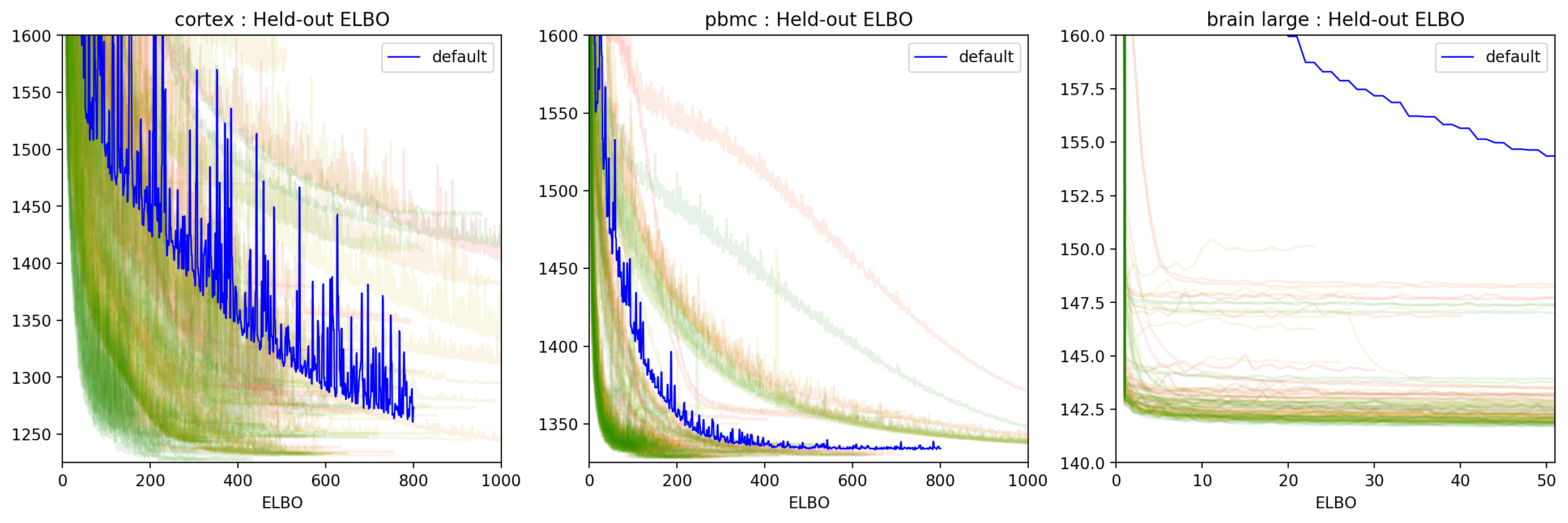
10.16. ELBO Histories plot¶
In the ELBO histories plotted below, the runs are colored from red to green, where red is the first run and green the last one.
[ ]:
plt.rcParams["figure.dpi"] = 200
plt.rcParams["figure.figsize"] = (10, 7)
[62]:
ylims_dict = {
"cortex": [1225, 1600],
"pbmc": [1325, 1600],
"brain large": [140, 160],
}
plotter.plot_histories(figsize=(17, 5), ylims_dict=ylims_dict, filename="elbo_histories_all", alpha=0.1)