CITE-seq analysis with totalVI#
With totalVI, we can produce a joint latent representation of cells, denoised data for both protein and RNA, integrate datasets, and compute differential expression of RNA and protein. Here we demonstrate this functionality with an integrated analysis of PBMC10k and PBMC5k, datasets of peripheral blood mononuclear cells publicly available from 10X Genomics subset to the 14 shared proteins between them. The same pipeline would generally be used to analyze a single CITE-seq dataset.
If you use totalVI, please consider citing:
Gayoso, A., Steier, Z., Lopez, R., Regier, J., Nazor, K. L., Streets, A., & Yosef, N. (2021). Joint probabilistic modeling of single-cell multi-omic data with totalVI. Nature Methods, 18(3), 272-282.
Note
Running the following cell will install tutorial dependencies on Google Colab only. It will have no effect on environments other than Google Colab.
!pip install --quiet scvi-colab
from scvi_colab import install
install()
WARNING: Running pip as the 'root' user can result in broken permissions and conflicting behaviour with the system package manager. It is recommended to use a virtual environment instead: https://pip.pypa.io/warnings/venv
import tempfile
import anndata as ad
import matplotlib.pyplot as plt
import mudata as md
import muon
import scanpy as sc
import scvi
import seaborn as sns
import torch
Imports and data loading#
scvi.settings.seed = 0
print("Last run with scvi-tools version:", scvi.__version__)
Last run with scvi-tools version: 1.1.0
Note
You can modify save_dir below to change where the data files for this tutorial are saved.
sc.set_figure_params(figsize=(6, 6), frameon=False)
sns.set_theme()
torch.set_float32_matmul_precision("high")
save_dir = tempfile.TemporaryDirectory()
%config InlineBackend.print_figure_kwargs={"facecolor": "w"}
%config InlineBackend.figure_format="retina"
This dataset was filtered as described in the totalVI manuscript (low quality cells, doublets, lowly expressed genes, etc.).
We run the standard workflow for keeping count and normalized data together.
adata = scvi.data.pbmcs_10x_cite_seq(save_path=save_dir.name)
adata
INFO Downloading file at /tmp/tmpf7863_f4/pbmc_10k_protein_v3.h5ad
Downloading...: 24938it [00:00, 87475.97it/s]
INFO Downloading file at /tmp/tmpf7863_f4/pbmc_5k_protein_v3.h5ad
Downloading...: 100%|██████████| 18295/18295.0 [00:00<00:00, 81284.01it/s]
AnnData object with n_obs × n_vars = 10849 × 15792
obs: 'n_genes', 'percent_mito', 'n_counts', 'batch'
obsm: 'protein_expression'
adata.layers["counts"] = adata.X.copy()
sc.pp.normalize_total(adata)
sc.pp.log1p(adata)
adata.obs_names_make_unique()
Important
In this tutorial we will show totalVI’s compatibility with the MuData format, which is a container for multiple AnnData objects. MuData objects can be read from the outputs of CellRanger using muon.read_10x_h5.
Furthermore, AnnData alone can also be used by storing the protein count data in .obsm, which is how it already is. For the AnnData-only workflow, see the documentation for setup_anndata in scvi.model.TOTALVI.
protein_adata = ad.AnnData(adata.obsm["protein_expression"])
protein_adata.obs_names = adata.obs_names
del adata.obsm["protein_expression"]
mdata = md.MuData({"rna": adata, "protein": protein_adata})
mdata
MuData object with n_obs × n_vars = 10849 × 15806
2 modalities
rna: 10849 x 15792
obs: 'n_genes', 'percent_mito', 'n_counts', 'batch'
uns: 'log1p'
layers: 'counts'
protein: 10849 x 14sc.pp.highly_variable_genes(
mdata.mod["rna"],
n_top_genes=4000,
flavor="seurat_v3",
batch_key="batch",
layer="counts",
)
# Place subsetted counts in a new modality
mdata.mod["rna_subset"] = mdata.mod["rna"][
:, mdata.mod["rna"].var["highly_variable"]
].copy()
mdata.update()
Setup mudata#
Now we run setup_mudata, which is the MuData analog to setup_anndata. The caveat of this workflow is that we need to provide this function which modality of the mdata object contains each piece of data. So for example, the batch information is in mdata.mod["rna"].obs["batch"]. Therefore, in the modalities argument below we specify that the batch_key can be found in the "rna_subset" modality of the MuData object.
Notably, we provide protein_layer=None. This means scvi-tools will pull information from .X from the modality specified in modalities ("protein" in this case). In the case of RNA, we want to use the counts, which we stored in mdata.mod["rna"].layers["counts"].
scvi.model.TOTALVI.setup_mudata(
mdata,
rna_layer="counts",
protein_layer=None,
batch_key="batch",
modalities={
"rna_layer": "rna_subset",
"protein_layer": "protein",
"batch_key": "rna_subset",
},
)
Note
Specify the modality of each argument via the modalities dictionary, which maps layer/key arguments to MuData modalities.
Prepare and run model#
model = scvi.model.TOTALVI(mdata)
INFO Computing empirical prior initialization for protein background.
model.train()
Epoch 287/400: 72%|███████▏ | 286/400 [01:54<00:45, 2.50it/s, v_num=1, train_loss_step=1.33e+3, train_loss_epoch=1.22e+3]Epoch 00287: reducing learning rate of group 0 to 2.4000e-03.
Epoch 351/400: 88%|████████▊ | 350/400 [02:19<00:18, 2.64it/s, v_num=1, train_loss_step=1.25e+3, train_loss_epoch=1.22e+3]Epoch 00351: reducing learning rate of group 0 to 1.4400e-03.
Epoch 365/400: 91%|█████████▏| 365/400 [02:24<00:13, 2.52it/s, v_num=1, train_loss_step=1.22e+3, train_loss_epoch=1.21e+3]
Monitored metric elbo_validation did not improve in the last 45 records. Best score: 1241.023. Signaling Trainer to stop.
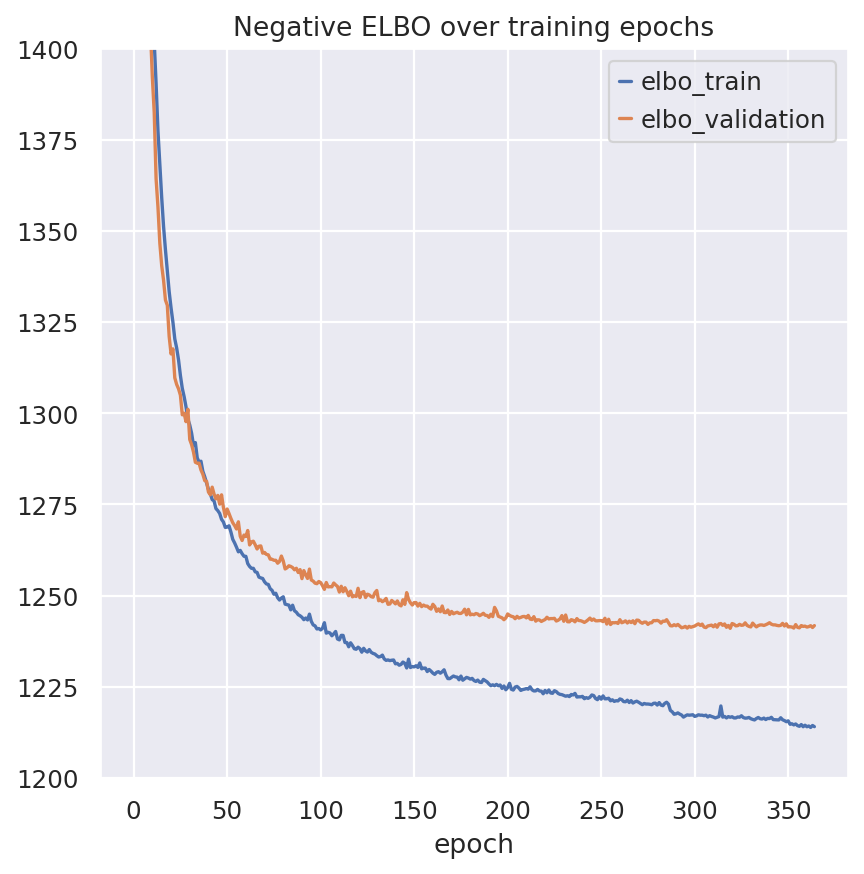
fig, ax = plt.subplots(1, 1)
model.history["elbo_train"].plot(ax=ax, label="train")
model.history["elbo_validation"].plot(ax=ax, label="validation")
ax.set(title="Negative ELBO over training epochs", ylim=(1200, 1400))
ax.legend()
<matplotlib.legend.Legend at 0x7f77359fa250>
Analyze outputs#
We use Scanpy and muon for clustering and visualization after running totalVI. It’s also possible to save totalVI outputs for an R-based workflow.
rna = mdata.mod["rna_subset"]
protein = mdata.mod["protein"]
# arbitrarily store latent in rna modality
TOTALVI_LATENT_KEY = "X_totalVI"
rna.obsm[TOTALVI_LATENT_KEY] = model.get_latent_representation()
rna_denoised, protein_denoised = model.get_normalized_expression(
n_samples=25, return_mean=True, transform_batch=["PBMC10k", "PBMC5k"]
)
rna.layers["denoised_rna"] = rna_denoised
protein.layers["denoised_protein"] = protein_denoised
protein.layers["protein_foreground_prob"] = (
100
* model.get_protein_foreground_probability(
n_samples=25, return_mean=True, transform_batch=["PBMC10k", "PBMC5k"]
)
)
parsed_protein_names = [p.split("_")[0] for p in protein.var_names]
protein.var["clean_names"] = parsed_protein_names
mdata.update()
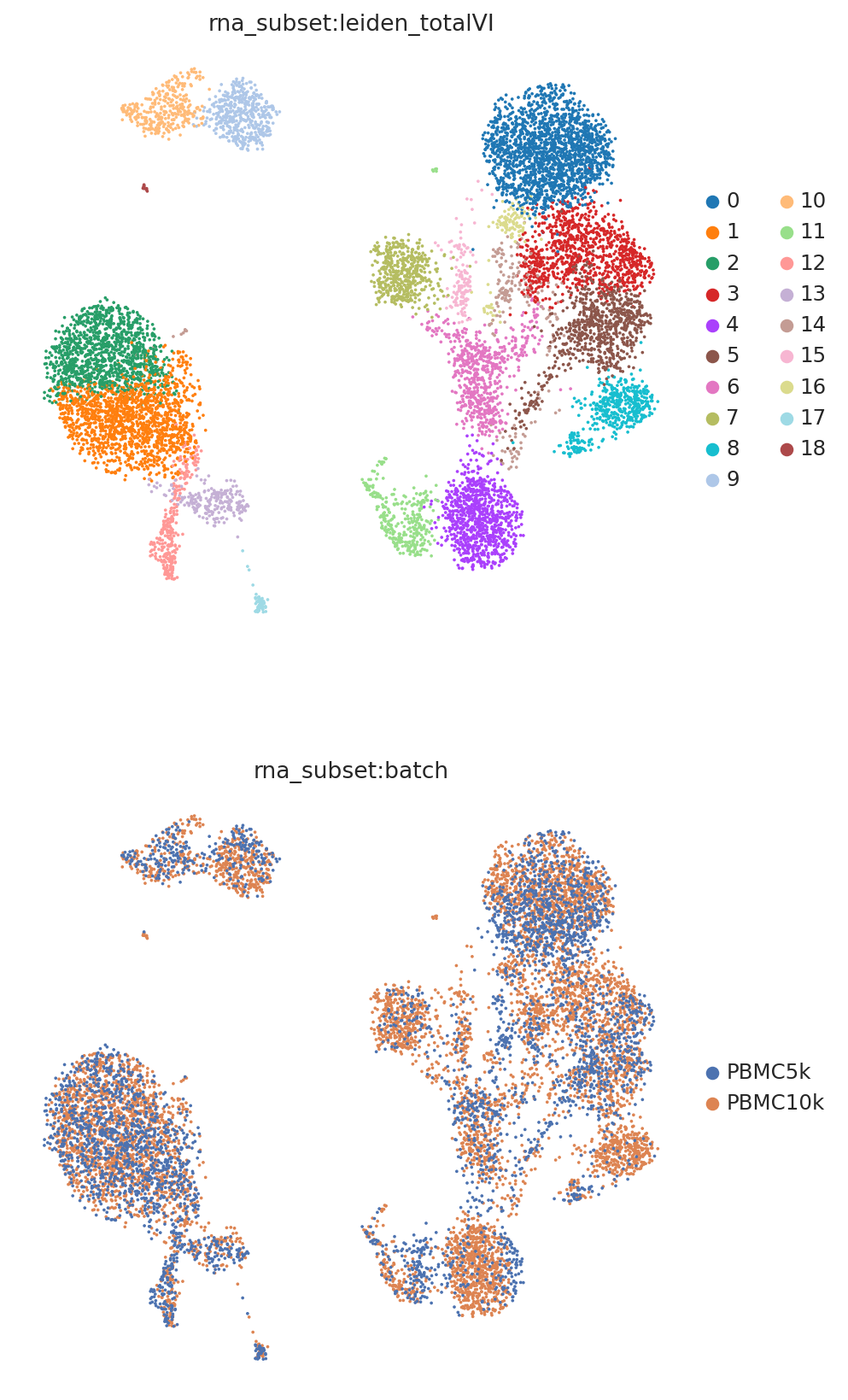
Now we can compute clusters and visualize the latent space.
TOTALVI_CLUSTERS_KEY = "leiden_totalVI"
sc.pp.neighbors(rna, use_rep=TOTALVI_LATENT_KEY)
sc.tl.umap(rna)
sc.tl.leiden(rna, key_added=TOTALVI_CLUSTERS_KEY)
mdata.update()
We can now use muon plotting functions which can pull data from either modality of the MuData object.
muon.pl.embedding(
mdata,
basis="rna_subset:X_umap",
color=[f"rna_subset:{TOTALVI_CLUSTERS_KEY}", "rna_subset:batch"],
frameon=False,
ncols=1,
)
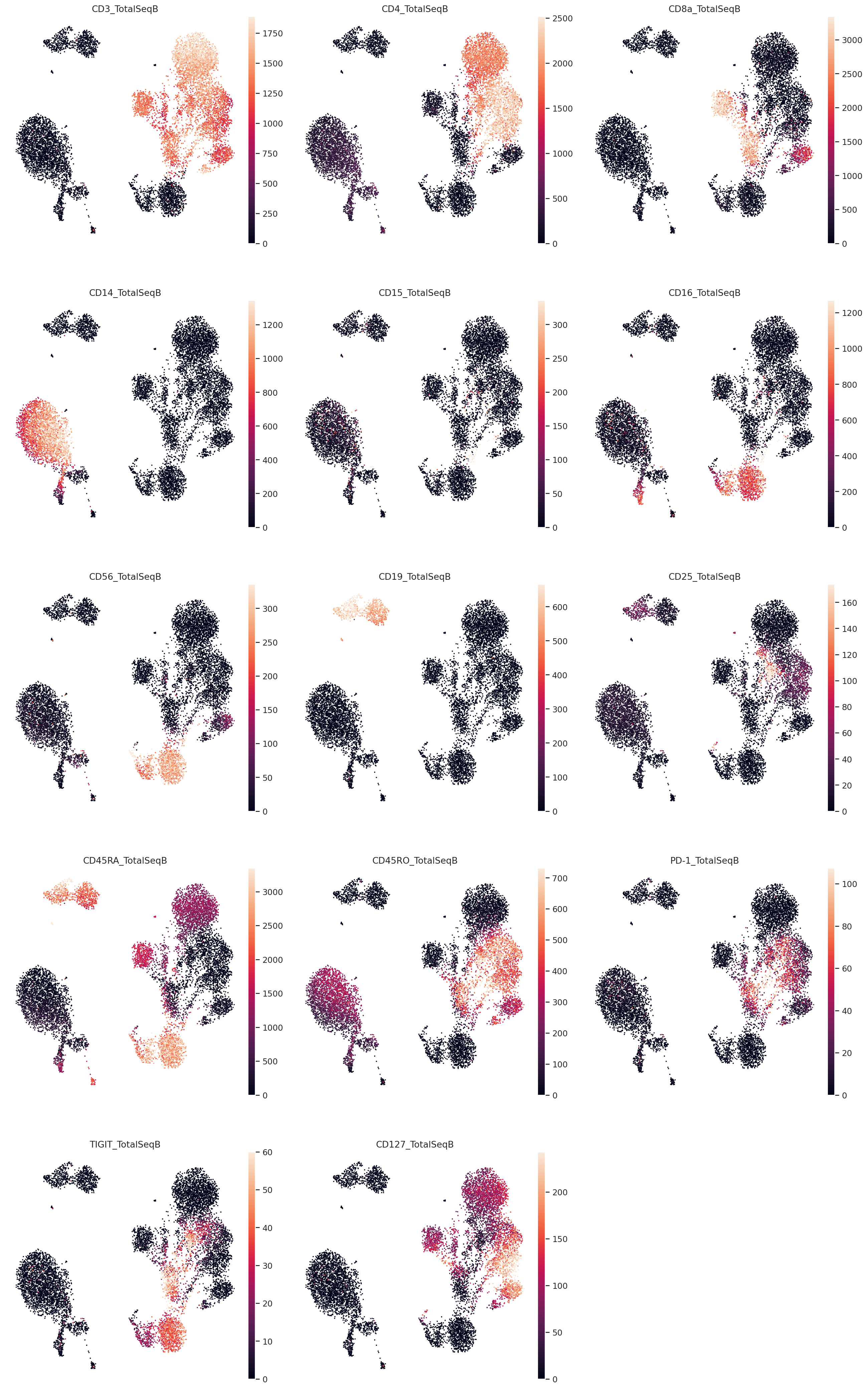
Visualize denoised protein values#
muon.pl.embedding(
mdata,
basis="rna_subset:X_umap",
color=protein.var_names,
frameon=False,
ncols=3,
vmax="p99",
wspace=0.1,
layer="denoised_protein",
)
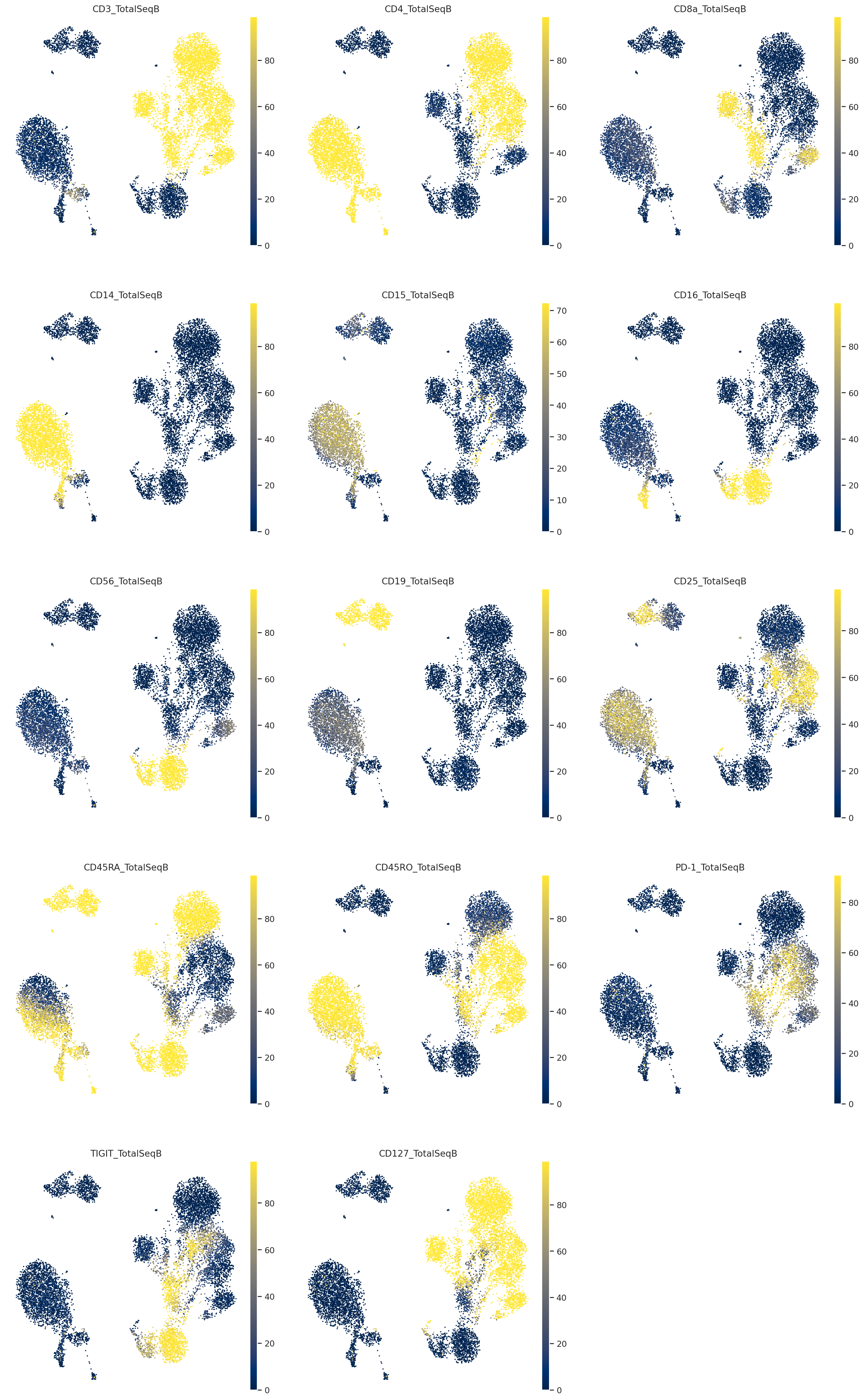
Visualize probability of foreground#
Here we visualize the probability of foreground for each protein and cell (projected on UMAP). Some proteins are easier to disentangle than others. Some proteins end up being “all background”. For example, CD15 does not appear to be captured well, when looking at the denoised values above we see little localization in the monocytes.
Note
While the foreground probability could theoretically be used to identify cell populations, we recommend using the denoised protein expression, which accounts for the foreground/background probability, but preserves the dynamic range of the protein measurements. Consequently, the denoised values are on the same scale as the raw data and it may be desirable to take a transformation like log or square root.
By viewing the foreground probability, we can get a feel for the types of cells in our dataset. For example, it’s very easy to see a population of monocytes based on the CD14 foregroud probability.
muon.pl.embedding(
mdata,
basis="rna_subset:X_umap",
layer="protein_foreground_prob",
color=protein.var_names,
frameon=False,
ncols=3,
vmax="p99",
wspace=0.1,
color_map="cividis",
)
Differential expression#
Here we do a one-vs-all DE test, where each cluster is tested against all cells not in that cluster. The results for each of the one-vs-all tests is concatenated into one DataFrame object. Inividual tests can be sliced using the “comparison” column. Genes and proteins are included in the same DataFrame.
Important
We do not recommend using totalVI denoised values in other differential expression tools, as denoised values are a summary of a random quantity. The totalVI DE test takes into account the full uncertainty of the denoised quantities.
de_df = model.differential_expression(
groupby="rna_subset:leiden_totalVI", delta=0.5, batch_correction=True
)
de_df.head(5)
DE...: 100%|██████████| 19/19 [00:17<00:00, 1.09it/s]
| proba_de | proba_not_de | bayes_factor | scale1 | scale2 | pseudocounts | delta | lfc_mean | lfc_median | lfc_std | ... | raw_mean1 | raw_mean2 | non_zeros_proportion1 | non_zeros_proportion2 | raw_normalized_mean1 | raw_normalized_mean2 | is_de_fdr_0.05 | comparison | group1 | group2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ANKRD55 | 0.9862 | 0.0138 | 4.269190 | 0.000099 | 0.000006 | 0.0 | 0.5 | 6.353043 | 6.681940 | 2.812859 | ... | 0.127775 | 0.009220 | 0.116405 | 0.008776 | 1.641991 | 0.088449 | True | 0 vs Rest | 0 | Rest |
| REG4 | 0.9812 | 0.0188 | 3.954919 | 0.000009 | 0.000004 | 0.0 | 0.5 | 5.717385 | 6.070202 | 3.558983 | ... | 0.008121 | 0.004110 | 0.007580 | 0.003777 | 0.086721 | 0.039889 | True | 0 vs Rest | 0 | Rest |
| MYO1F | 0.9776 | 0.0224 | 3.776039 | 0.000035 | 0.000744 | 0.0 | 0.5 | -4.554471 | -4.926318 | 2.183192 | ... | 0.013535 | 1.470562 | 0.012453 | 0.589425 | 0.171044 | 7.967407 | True | 0 vs Rest | 0 | Rest |
| CA6 | 0.9760 | 0.0240 | 3.705408 | 0.000010 | 0.000002 | 0.0 | 0.5 | 5.712860 | 5.919196 | 3.640816 | ... | 0.010287 | 0.002777 | 0.010287 | 0.002222 | 0.109388 | 0.027540 | True | 0 vs Rest | 0 | Rest |
| NOG | 0.9756 | 0.0244 | 3.688469 | 0.000244 | 0.000016 | 0.0 | 0.5 | 8.877318 | 9.526344 | 3.827954 | ... | 0.217109 | 0.016219 | 0.178127 | 0.014219 | 2.714708 | 0.207277 | True | 0 vs Rest | 0 | Rest |
5 rows × 22 columns
Now we filter the results such that we retain features above a certain Bayes factor (which here is on the natural log scale) and genes with greater than 10% non-zero entries in the cluster of interest.
filtered_pro = {}
filtered_rna = {}
cats = rna.obs[TOTALVI_CLUSTERS_KEY].cat.categories
for i, c in enumerate(cats):
cid = f"{c} vs Rest"
cell_type_df = de_df.loc[de_df.comparison == cid]
cell_type_df = cell_type_df.sort_values("lfc_median", ascending=False)
cell_type_df = cell_type_df[cell_type_df.lfc_median > 0]
pro_rows = cell_type_df.index.str.contains("TotalSeqB")
data_pro = cell_type_df.iloc[pro_rows]
data_pro = data_pro[data_pro["bayes_factor"] > 0.7]
data_rna = cell_type_df.iloc[~pro_rows]
data_rna = data_rna[data_rna["bayes_factor"] > 3]
data_rna = data_rna[data_rna["non_zeros_proportion1"] > 0.1]
filtered_pro[c] = data_pro.index.tolist()[:3]
filtered_rna[c] = data_rna.index.tolist()[:2]
We can also use general scanpy visualization functions
sc.tl.dendrogram(rna, groupby=TOTALVI_CLUSTERS_KEY, use_rep=TOTALVI_LATENT_KEY)
# This is a bit of a hack to be able to use scanpy dendrogram with the protein data
protein.obs[TOTALVI_CLUSTERS_KEY] = rna.obs[TOTALVI_CLUSTERS_KEY]
protein.obsm[TOTALVI_LATENT_KEY] = rna.obsm[TOTALVI_LATENT_KEY]
sc.tl.dendrogram(protein, groupby=TOTALVI_CLUSTERS_KEY, use_rep=TOTALVI_LATENT_KEY)
sc.pl.dotplot(
rna,
filtered_rna,
groupby=TOTALVI_CLUSTERS_KEY,
dendrogram=True,
standard_scale="var",
swap_axes=True,
)
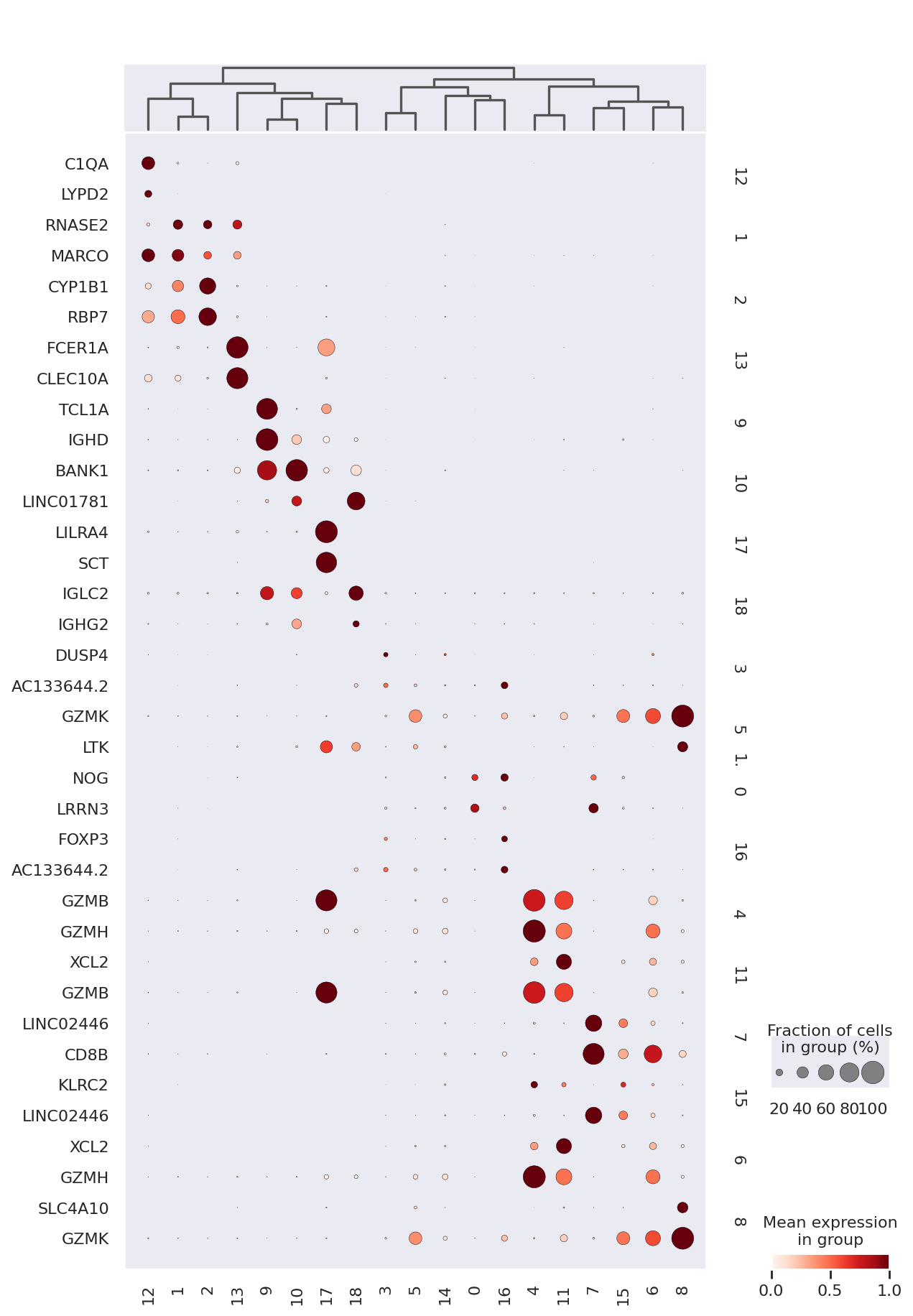
Matrix plot displays totalVI denoised protein expression per leiden cluster.
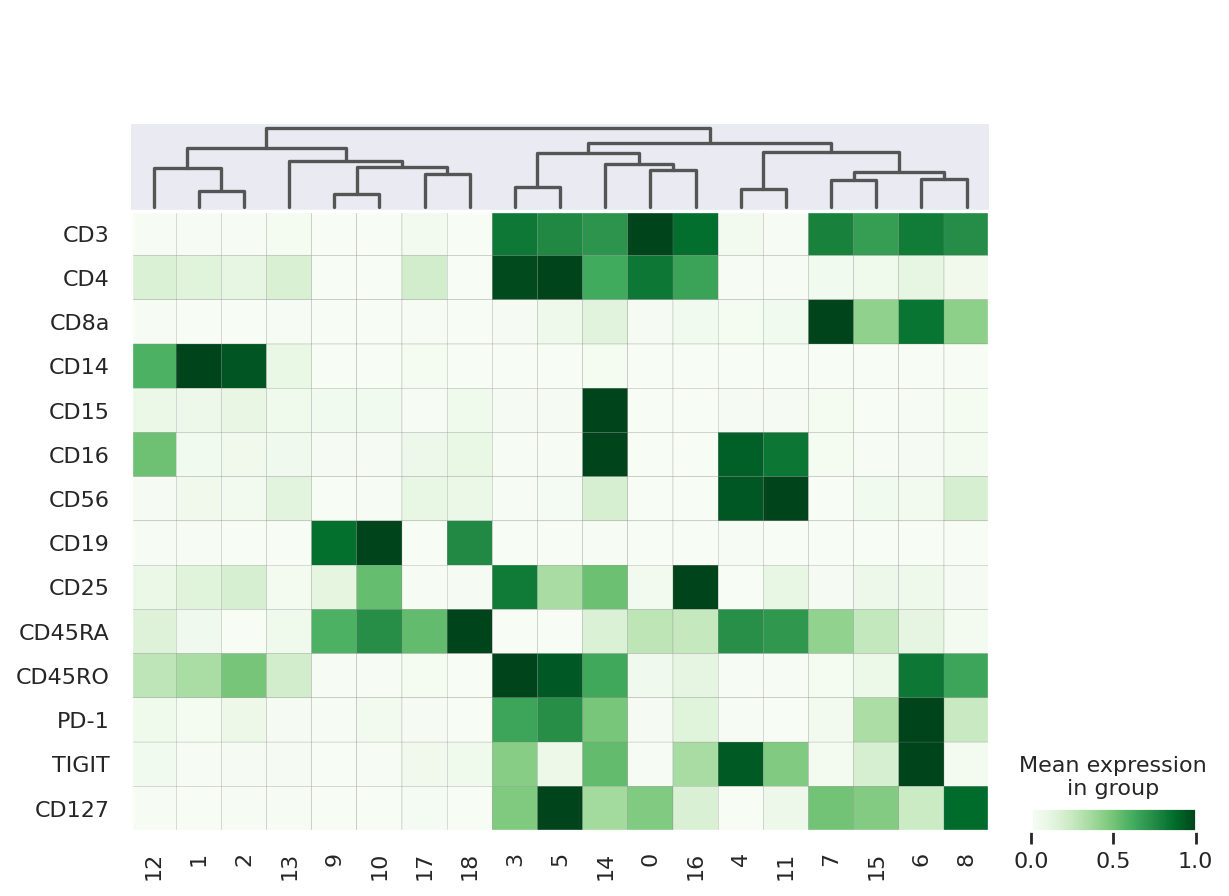
sc.pl.matrixplot(
protein,
protein.var["clean_names"],
groupby=TOTALVI_CLUSTERS_KEY,
gene_symbols="clean_names",
dendrogram=True,
swap_axes=True,
layer="denoised_protein",
cmap="Greens",
standard_scale="var",
)
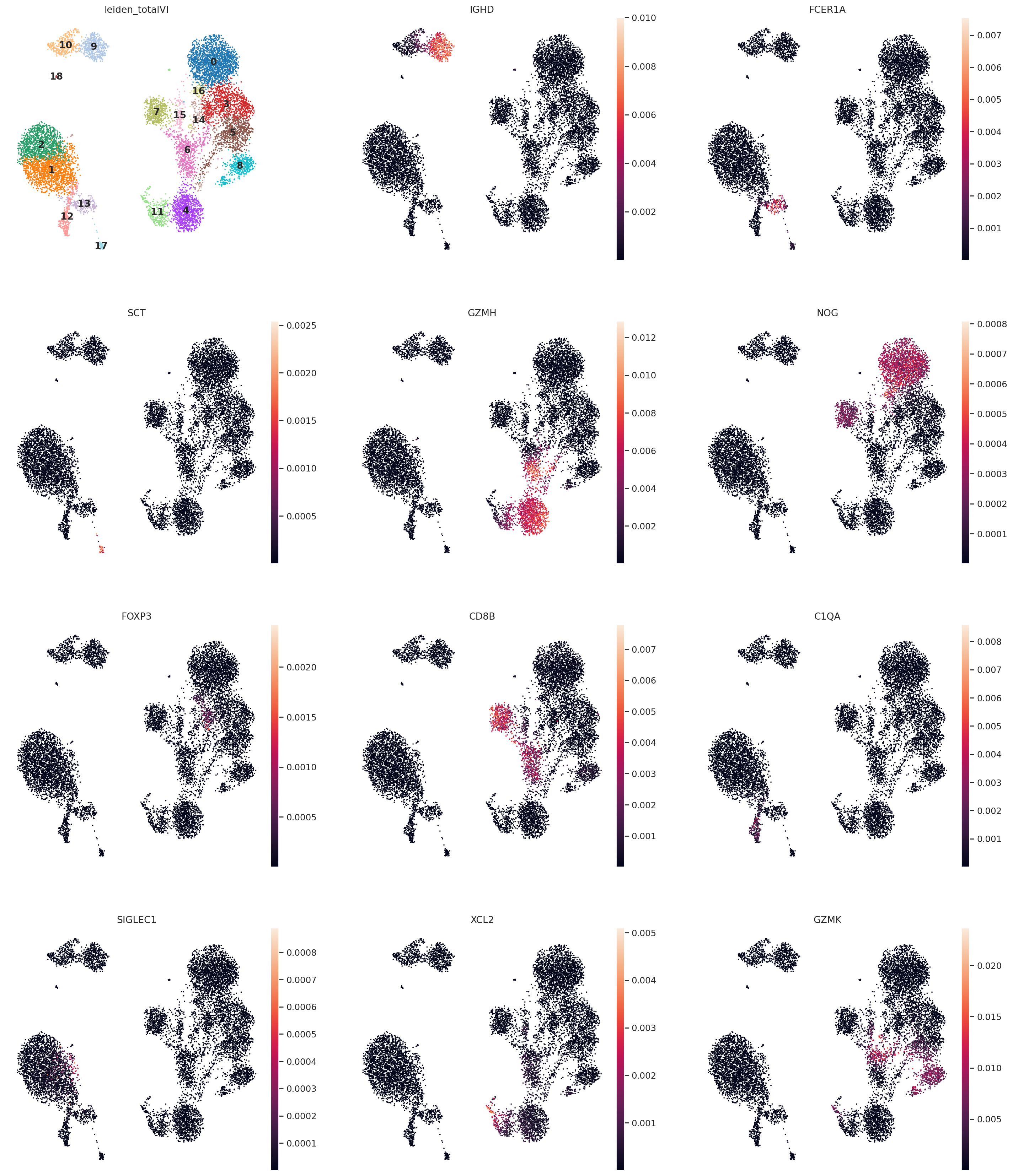
This is a selection of some of the markers that turned up in the RNA DE test.
sc.pl.umap(
rna,
color=[
TOTALVI_CLUSTERS_KEY,
"IGHD",
"FCER1A",
"SCT",
"GZMH",
"NOG",
"FOXP3",
"CD8B",
"C1QA",
"SIGLEC1",
"XCL2",
"GZMK",
],
legend_loc="on data",
frameon=False,
ncols=3,
layer="denoised_rna",
wspace=0.2,
)