User guide#
scvi-tools is composed of models that can perform one or many analysis tasks. In the user guide, we provide an overview of each model with emphasis on the math behind the model, how it connects to the code, and how the code connects to analysis.
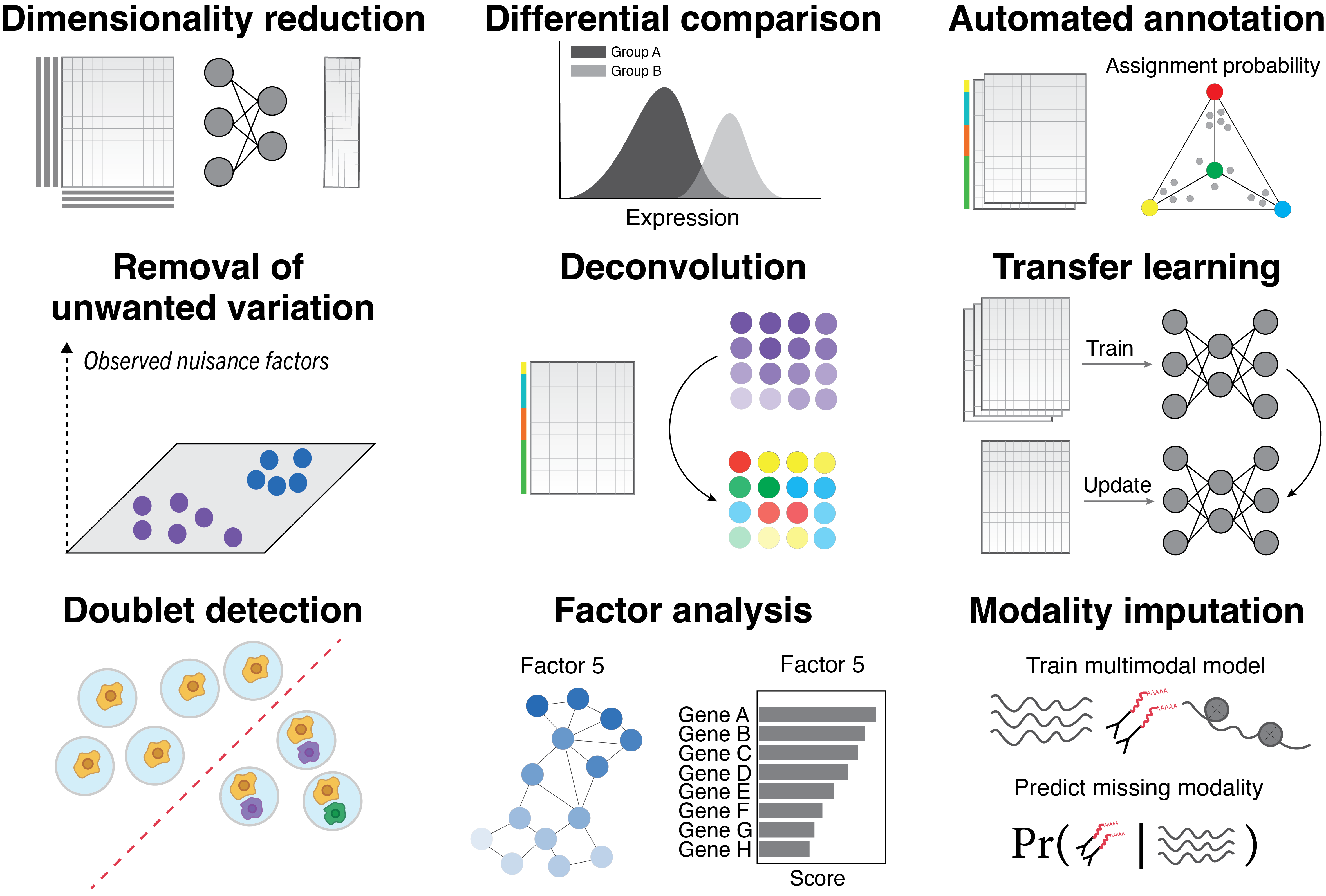
scRNA-seq analysis#
Model |
Tasks |
Reference |
|---|---|---|
Dimensionality reduction, removal of unwanted variation, integration across replicates, donors, and technologies, differential expression, imputation, normalization of other cell- and sample-level confounding factors |
||
scVI tasks with cell type transfer from reference, seed labeling |
||
scVI tasks with linear decoder |
||
for assessing gene-specific levels of zero-inflation in scRNA-seq data |
||
Marker-based automated annotation |
||
Doublet detection |
||
Ambient RNA removal |
||
scVI tasks with contrastive analysis |
ATAC-seq analysis#
Model |
Tasks |
Reference |
|---|---|---|
Dimensionality reduction, removal of unwanted variation, integration across replicates, donors, and technologies, differential expression, imputation, normalization of other cell- and sample-level confounding factors |
||
Dimensionality reduction, removal of unwanted variation, integration across replicates, donors, and technologies, imputation |
||
Dimensionality reduction, removal of unwanted variation, integration across replicates, donors, and technologies, differential expression, imputation, normalization of other cell- and sample-level confounding factors |
Multimodal analysis#
CITE-seq#
Model |
Tasks |
Reference |
|---|---|---|
Dimensionality reduction, removal of unwanted variation, integration across replicates, donors, and technologies, differential expression, protein imputation, imputation, normalization of other cell- and sample-level confounding factors |
Multiome#
Model |
Tasks |
Reference |
|---|---|---|
Integration of paired/unpaired multiome data, missing modality imputation, normalization of other cell- and sample-level confounding factors |
Spatial transcriptomics analysis#
Model |
Tasks |
Reference |
|---|---|---|
Multi-resolution deconvolution, cell-type-specific gene expression imputation, comparative analysis |
||
Deconvolution |
||
Imputation of missing spatial genes |
||
Deconvolution, single cell spatial mapping |
General purpose analysis#
Model |
Tasks |
Reference |
|---|---|---|
Topic modeling |
Background#
Glossary#
A Model class inherits BaseModelClass and is the user-facing object for interacting with a module.
The model has a train method that learns the parameters of the module, and also contains methods
for users to retrieve information from the module, like the latent representation of cells in a VAE.
Conventionally, the post-inference model methods should not store data into the AnnData object, but
instead return “standard” Python objects, like numpy arrays or pandas dataframes.
A module is the lower-level object that defines a generative model and inference scheme. A module will
either inherit BaseModuleClass or PyroBaseModuleClass.
Consequently, a module can either be implemented with PyTorch alone, or Pyro. In the PyTorch only case, the
generative process and inference scheme are implemented respectively in the generative and inference methods,
while the loss method computes the loss, e.g, ELBO in the case of variational inference.
The training plan is a PyTorch Lightning Module that is initialized with a scvi-tools module object. It configures the optimizers, defines the training step and validation step, and computes metrics to be recorded during training. The training step and validation step are functions that take data, run it through the model and return the loss, which will then be used to optimize the model parameters in the Trainer. Overall, custom training plans can be used to develop complex inference schemes on top of modules.
The Trainer is a lightweight wrapper of the PyTorch Lightning Trainer. It takes as input
the training plan, a training data loader, and a validation dataloader. It performs the actual training loop, in
which parameters are optimized, as well as the validation loop to monitor metrics. It automatically handles moving
data to the correct device (CPU/GPU).
