Differential expression on C. elegans data#
This notebook was contributed by Eduardo Beltrame @Munfred and edited by Romain Lopez, Adam Gayoso, and Pierre Boyeau.
Processing and visualizing 89k cells from Packer et al. 2019 C. elegans 10xv2 single cell data
Original article:
A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution
https://science.sciencemag.org/content/365/6459/eaax1971.long
The anndata object we provide has 89,701 cells and 20,222 genes. It includes short gene descriptions from WormBase that will show up when mousing over the interactive plots.
Steps performed:
Loading the data from anndata containing cell labels and gene descriptions
Training the model with batch labels for integration with scVI
Retrieving the scVI latent space and imputed values
Visualize the latent space with an interactive t-SNE plot using Plotly
Perform differential expression and visualize with interactive volcano plot and heatmap using Plotly
This notebook was designed to be run in Google Colab.
Note
Running the following cell will install tutorial dependencies on Google Colab only. It will have no effect on environments other than Google Colab.
!pip install --quiet scvi-colab
from scvi_colab import install
install()
WARNING: Running pip as the 'root' user can result in broken permissions and conflicting behaviour with the system package manager. It is recommended to use a virtual environment instead: https://pip.pypa.io/warnings/venv
import os
import tempfile
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import plotnine as p9
import scanpy as sc
import scvi
import seaborn as sns
import torch
scvi.settings.seed = 0
print("Last run with scvi-tools version:", scvi.__version__)
Last run with scvi-tools version: 1.1.0
Note
You can modify save_dir below to change where the data files for this tutorial are saved.
sc.set_figure_params(figsize=(6, 6), frameon=False)
sns.set_theme()
torch.set_float32_matmul_precision("high")
save_dir = tempfile.TemporaryDirectory()
%config InlineBackend.print_figure_kwargs={"facecolor": "w"}
%config InlineBackend.figure_format="retina"
adata_path = os.path.join(save_dir.name, "packer2019.h5ad")
adata = sc.read(
adata_path,
backup_url="https://github.com/Munfred/wormcells-site/releases/download/packer2019/packer2019.h5ad",
)
adata
AnnData object with n_obs × n_vars = 89701 × 20222
obs: 'cell', 'numi', 'time_point', 'batch', 'size_factor', 'cell_type', 'cell_subtype', 'plot_cell_type', 'raw_embryo_time', 'embryo_time', 'embryo_time_bin', 'raw_embryo_time_bin', 'lineage', 'passed_qc'
var: 'gene_id', 'gene_name', 'gene_description'
Take a look at the gene descriptions#
The gene descriptions were taken using the WormBase API.
display(
adata.var.head().style.set_properties(
subset=["gene_description"], **{"width": "600px"}
)
)
| gene_id | gene_name | gene_description | |
|---|---|---|---|
| index | |||
| WBGene00010957 | WBGene00010957 | nduo-6 | Is affected by several genes including daf-16; daf-12; and hsf-1 based on RNA-seq and tiling array studies. Is affected by six chemicals including Rotenone; Psoralens; and metformin based on RNA-seq and microarray studies. |
| WBGene00010958 | WBGene00010958 | ndfl-4 | Is enriched in Psub2 based on RNA-seq studies. Is affected by several genes including daf-16; daf-12; and clk-1 based on RNA-seq and microarray studies. Is affected by six chemicals including Alovudine; Psoralens; and metformin based on RNA-seq studies. |
| WBGene00010959 | WBGene00010959 | nduo-1 | Is an ortholog of human MT-ND1 (mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 1). Is predicted to contribute to NADH dehydrogenase activity. Human ortholog(s) of this gene are implicated in Leber hereditary optic neuropathy and MELAS syndrome. |
| WBGene00010960 | WBGene00010960 | atp-6 | Is predicted to contribute to proton-transporting ATP synthase activity, rotational mechanism. |
| WBGene00010961 | WBGene00010961 | nduo-2 | Is affected by several genes including hsf-1; clk-1; and elt-2 based on RNA-seq and microarray studies. Is affected by eight chemicals including stavudine; Zidovudine; and Psoralens based on RNA-seq and microarray studies. |
Selecting genes and loading data#
We use the utility scvi.data.poisson_gene_selection to select genes according to their dropout rate, which is a simple and scalable approach to select genes.
This method was described by Andrews & Hemberg in the article M3Drop: dropout-based feature selection for scRNASeq :
https://academic.oup.com/bioinformatics/article/35/16/2865/5258099
This method modifies the adata to add the following fields:
highly_variable # boolean true for chosen genes
observed_fraction_zeros # fraction of observed zeros per gene
expected_fraction_zeros # expected fraction of observed zeros per gene
prob_zero_enriched_nbatches # If batch_key is given, this denotes in how many batches genes are detected as zero enriched
prob_zero_enrichment # Probability of zero enrichment, median across batches in the case of multiple batches
prob_zero_enrichment_rank # Rank of the gene according to probability of zero enrichment
Note
Gene selection is an important step to obtain relevant cell representations with scVI. Generally, selecting the top few thousands top-ranking genes predicted by a gene selection tool suffice to obtain good performance.
Increasing the number of selected genes may be required in some applications, e.g., to increase the number of considered genes for differential expression. Note, however, that this will increase the time required to reach convergence and GPU memory load. It may also require to tune scVI’s model hyperparameters (see the autotune tutorial)
Alternatives to the Poisson gene selection can be used, e.g., via scanpy or seurat.
scvi.data.poisson_gene_selection(adata)
adata.var.head()
Sampling from binomial...: 100%|██████████| 10000/10000 [00:00<00:00, 21246.35it/s]
| gene_id | gene_name | gene_description | highly_variable | observed_fraction_zeros | expected_fraction_zeros | prob_zero_enriched_nbatches | prob_zero_enrichment | prob_zero_enrichment_rank | |
|---|---|---|---|---|---|---|---|---|---|
| index | |||||||||
| WBGene00010957 | WBGene00010957 | nduo-6 | Is affected by several genes including daf-16;... | False | 0.075685 | 0.002028 | 0 | 0.0762 | 15661.0 |
| WBGene00010958 | WBGene00010958 | ndfl-4 | Is enriched in Psub2 based on RNA-seq studies.... | True | 0.659680 | 0.623356 | 1 | 0.2502 | 19567.0 |
| WBGene00010959 | WBGene00010959 | nduo-1 | Is an ortholog of human MT-ND1 (mitochondriall... | True | 0.201993 | 0.046608 | 1 | 0.1886 | 18907.0 |
| WBGene00010960 | WBGene00010960 | atp-6 | Is predicted to contribute to proton-transport... | True | 0.138081 | 0.001848 | 1 | 0.1381 | 17859.0 |
| WBGene00010961 | WBGene00010961 | nduo-2 | Is affected by several genes including hsf-1; ... | True | 0.468434 | 0.383116 | 1 | 0.2860 | 19800.0 |
adata = adata[:, adata.var["highly_variable"]] # focus on selected genes
adata.layers[
"counts"
] = adata.X.copy().tocsr() # converts to CSR format, preserve counts
scvi.model.SCVI.setup_anndata(
adata, layer="counts", batch_key="batch"
) # prepare data for scVI
Define and train the model#
model = scvi.model.SCVI(
adata, gene_likelihood="nb"
) # We use Negative Binomial count likelihoods, following Boyeau et al., 2023.
model.train(
check_val_every_n_epoch=1,
max_epochs=400,
early_stopping=True,
early_stopping_patience=20,
early_stopping_monitor="elbo_validation",
)
Epoch 400/400: 100%|██████████| 400/400 [20:15<00:00, 3.04s/it, v_num=1, train_loss_step=2.03e+3, train_loss_epoch=2.06e+3]
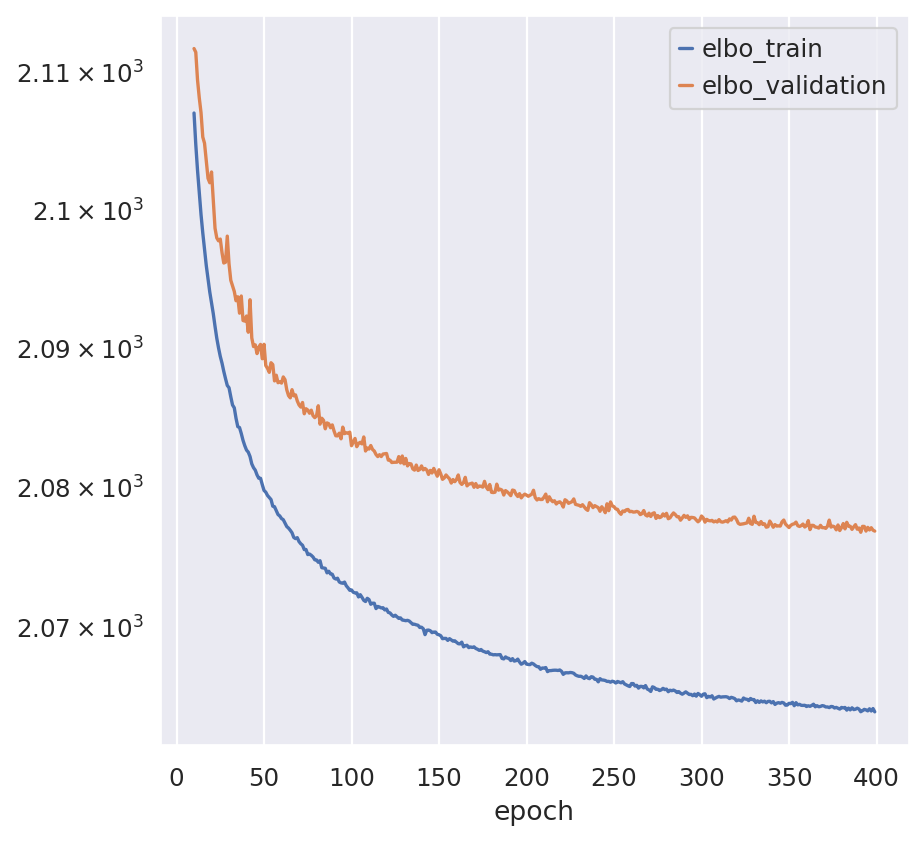
# Ensure convergence
train_test_results = model.history["elbo_train"]
train_test_results["elbo_validation"] = model.history["elbo_validation"]
train_test_results.iloc[10:].plot(logy=True) # exclude first 10 epochs
plt.show()
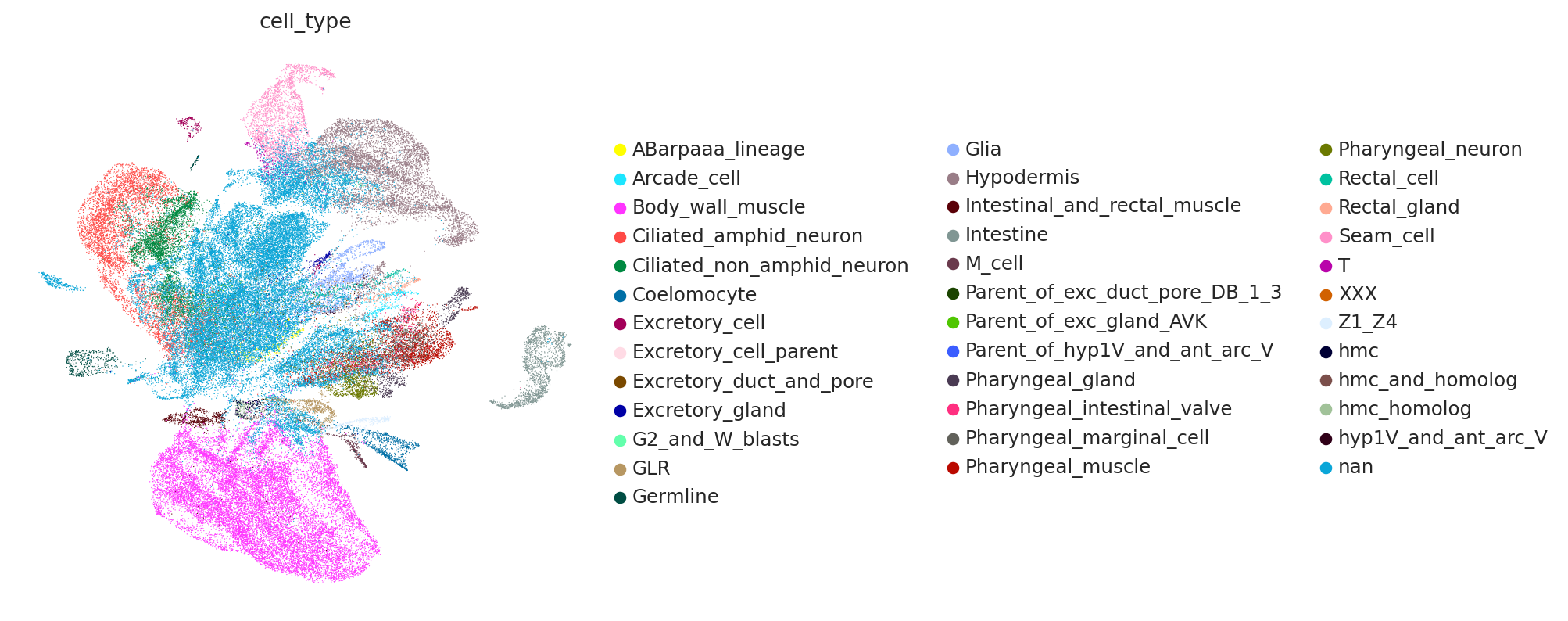
Get the latent space and compute UMAP#
SCVI_LATENT_KEY = "X_scVI"
latent = model.get_latent_representation()
adata.obsm[SCVI_LATENT_KEY] = latent
sc.pp.neighbors(adata, use_rep=SCVI_LATENT_KEY)
sc.tl.umap(adata)
sc.pl.umap(adata, color="cell_type")
Performing Differential Expression in scVI#
Differential expression (DE) analysis is used to quantify the differences in gene expression across subpopulations of genes. If we have two sets of cells \(A\) and \(B\), a DE test is typically used to predict two things for each gene:
Whether the gene’s expression levels are significantly different in the A and B sets of cells.
An effect size that quantifies the strength of the differential expression.
Once trained, scVI can natively perform both of these tasks. Additionally, its differential expression module can account for batch effects and filter DE genes expected to be of little relevance.
Selecting cell subpopulations to compare#
# let's take a look at abundances of different cell types
adata.obs["cell_type"].value_counts()
cell_type
nan 35052
Body_wall_muscle 17520
Hypodermis 7746
Ciliated_amphid_neuron 6090
Ciliated_non_amphid_neuron 4468
Seam_cell 2766
Pharyngeal_muscle 2562
Glia 1857
Intestine 1732
Pharyngeal_neuron 1471
Pharyngeal_marginal_cell 911
Coelomocyte 787
Pharyngeal_gland 786
GLR 768
Intestinal_and_rectal_muscle 568
Germline 499
Pharyngeal_intestinal_valve 493
Arcade_cell 434
Z1_Z4 372
Rectal_cell 327
M_cell 315
ABarpaaa_lineage 273
Rectal_gland 265
Excretory_cell 215
Excretory_gland 205
hmc 189
hmc_homolog 155
T 141
hmc_and_homolog 122
Parent_of_exc_gland_AVK 114
hyp1V_and_ant_arc_V 112
Excretory_duct_and_pore 91
Parent_of_hyp1V_and_ant_arc_V 75
G2_and_W_blasts 72
Excretory_cell_parent 62
Parent_of_exc_duct_pore_DB_1_3 61
XXX 25
Name: count, dtype: int64
scVI provides several options to identify the two populations of interest.
cell_type_1 = "Ciliated_non_amphid_neuron"
cell_idx1 = adata.obs["cell_type"] == cell_type_1
print(sum(cell_idx1), "cells of type", cell_type_1)
cell_type_2 = "Intestine"
cell_idx2 = adata.obs["cell_type"] == cell_type_2
print(sum(cell_idx2), "cells of type", cell_type_2)
# or equivalently, provide a string of the form "my_celltype_column == 'desired_celltype'"
# cell_idx1 = "cell_type == 'Ciliated_non_amphid_neuron'"
# cell_idx2 = "cell_type == 'Intestine'"
4468 cells of type Ciliated_non_amphid_neuron
1732 cells of type Intestine
Basic principle#
DE and log fold-changes#
scVI can natively be used to perform differential expression analyses to compare populations of cells, as described here and there. It achieves this by estimating the posterior distribution of the log fold-change (LFC) between subpopulations \(A\) and \(B\). Specifically, for a given gene g, scVI calculates the LFC as the difference between the logarithm of its expression level in population A, denoted by \(h_g^A\), and the logarithm of its expression level in population B, denoted by \(h_g^B\).
The resulting value, denoted by \(\beta_g\), provides insights into the expression patterns of gene \(g\). Values close to zero indicate that the gene is expressed similarly in both populations, positive values suggest upregulation in population A and negative values indicate downregulation in population A. This information can be used to better understand the biological mechanisms underlying the differences between the two cell populations.
DE testing#
In addition to estimating the LFC, scVI can also detect which genes have significant expression patterns. To tag which genes are differentially expressed, scVI tests the following competing hypotheses \(M_{1, g}: \beta_g \in [-\delta, \delta]\) and \(M_{2, g}: \beta_g \in (-\infty, -\delta) \cup (\delta, \infty)\). Here, \(\delta\) denotes a small LFC threshold, such that \(\beta_g \in [-\delta, \delta]\) is evidence that the gene is equally expressed in the two subpopulations.
Differentially expressed genes are identified by computing the posterior probability of \(M_{2, g}\).
Running DE analyses#
Running and understanding a DE run#
A simple DE analysis can then be performed using the following command
de_change = model.differential_expression(idx1=cell_idx1, idx2=cell_idx2)
de_change
DE...: 100%|██████████| 1/1 [00:00<00:00, 1.20it/s]
| proba_de | proba_not_de | bayes_factor | scale1 | scale2 | pseudocounts | delta | lfc_mean | lfc_median | lfc_std | lfc_min | lfc_max | raw_mean1 | raw_mean2 | non_zeros_proportion1 | non_zeros_proportion2 | raw_normalized_mean1 | raw_normalized_mean2 | is_de_fdr_0.05 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| index | |||||||||||||||||||
| WBGene00006870 | 1.0000 | 0.0000 | 18.420681 | 0.000400 | 0.000004 | 0.0 | 0.25 | 7.165903 | 7.197666 | 1.520534 | -1.765389 | 12.305556 | 0.567147 | 0.028291 | 0.252686 | 0.025982 | 5.500196 | 0.044442 | True |
| WBGene00011938 | 1.0000 | 0.0000 | 18.420681 | 0.000013 | 0.000480 | 0.0 | 0.25 | -5.446735 | -5.540369 | 1.405468 | -9.422789 | 1.125211 | 0.011638 | 3.357980 | 0.010072 | 0.834873 | 0.128841 | 4.526570 | True |
| WBGene00013560 | 0.9998 | 0.0002 | 8.516943 | 0.000038 | 0.001268 | 0.0 | 0.25 | -4.677629 | -4.729869 | 1.584986 | -9.786007 | 2.483212 | 0.035586 | 8.210736 | 0.030886 | 0.898961 | 0.340279 | 12.123684 | True |
| WBGene00011121 | 0.9998 | 0.0002 | 8.516943 | 0.001431 | 0.000006 | 0.0 | 0.25 | 7.113941 | 7.609168 | 2.303849 | -1.747072 | 12.854574 | 1.442006 | 0.040416 | 0.537601 | 0.038684 | 14.994382 | 0.059873 | True |
| WBGene00000452 | 0.9998 | 0.0002 | 8.516943 | 0.000240 | 0.000005 | 0.0 | 0.25 | 5.858909 | 5.913099 | 1.573072 | -1.599674 | 11.946387 | 0.376680 | 0.033487 | 0.123545 | 0.027136 | 3.191612 | 0.074427 | True |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| WBGene00022044 | 0.5808 | 0.4192 | 0.326058 | 0.000182 | 0.000167 | 0.0 | 0.25 | 0.164114 | 0.127701 | 0.512683 | -1.325256 | 3.128280 | 0.189124 | 1.164550 | 0.163608 | 0.618938 | 1.599760 | 1.586375 | False |
| WBGene00044608 | 0.5746 | 0.4254 | 0.300644 | 0.000109 | 0.000094 | 0.0 | 0.25 | 0.215397 | 0.201292 | 0.418493 | -1.350334 | 2.464341 | 0.146599 | 0.693418 | 0.123545 | 0.452079 | 1.315090 | 0.948504 | False |
| WBGene00022702 | 0.5600 | 0.4400 | 0.241162 | 0.000062 | 0.000062 | 0.0 | 0.25 | 0.000142 | -0.035827 | 0.532482 | -1.729041 | 3.303222 | 0.075649 | 0.457272 | 0.068039 | 0.340647 | 0.639998 | 0.621169 | False |
| WBGene00011116 | 0.5184 | 0.4816 | 0.073633 | 0.000302 | 0.000267 | 0.0 | 0.25 | 0.175932 | 0.171164 | 0.354389 | -1.125929 | 1.881635 | 0.356761 | 1.845267 | 0.275067 | 0.763279 | 2.979073 | 2.603784 | False |
| WBGene00011859 | 0.4998 | 0.5002 | -0.000800 | 0.000122 | 0.000115 | 0.0 | 0.25 | 0.099528 | 0.064282 | 0.494328 | -1.683570 | 3.587107 | 0.141899 | 0.824482 | 0.122426 | 0.515589 | 1.178068 | 1.153656 | False |
4000 rows × 19 columns
This method returns a pandas DataFrame, where each row corresponds to a gene.
The most important columns of this dataframe are the following.
proba_de, which captures the posterior probability of \(M_{2, g}\) that the gene is differentially expressed. Values close to one indicate that the gene is DE;
lfc_mean and lfc_median, respectively denoting the mean and the median of the posterior distribution of \(\beta_g\). Positive values of the LFC signify that the gene is upregulated in idx1;
is_de_fdr_0.05 is True when the gene is tagged DE after FDR correction at target level \(\alpha=0.05\). The target level can be adjusted by specifying fdr_target in the differential_expression method.
Main hyperparameters#
Additional parameters can be tuned in specific applications.
weights, which can correspond to ”uniform” or ”importance” specify how normalized gene expressions in the two subpopulations are computed. The ”importance” flavor is specifically designed to provide FDR-calibrated gene sets, but may be overkill if you only aim to rank genes (via their LFC for instance).
filter_outlier_cells filters out outlier cells prior to computing normalized gene expressions. It is important to set this parameter to
Truewhenweights='importance'.delta allows to specify \(\delta\), which is used to detect differentially expressed genes. It can also be set automatically using
delta=Nonepseudocounts, which is zero by default, but can be set to a small value (e.g., 1e-6) when many detected DE genes are seldom expressed in the compared populations.
batch_correction, which should be set to
Trueto account for batch effects. This only makes sense whenidx1andidx2denote cells coming from overlapping batches.
The exact function of these parameters is described here.
Volcano plot of change mode DE with p-values#
de_change_uniform = model.differential_expression(
idx1=cell_idx1, # we use the same cells as chosen before
idx2=cell_idx2,
weights="uniform",
batch_correction=True,
)
# manipulate the DE results for plotting
de_change_uniform["log10_pscore"] = np.log10(de_change_uniform["proba_not_de"])
de_change_uniform = de_change_uniform.join(adata.var, how="inner")
de_change_uniform.head()
DE...: 100%|██████████| 1/1 [00:01<00:00, 1.54s/it]
| proba_de | proba_not_de | bayes_factor | scale1 | scale2 | pseudocounts | delta | lfc_mean | lfc_median | lfc_std | ... | log10_pscore | gene_id | gene_name | gene_description | highly_variable | observed_fraction_zeros | expected_fraction_zeros | prob_zero_enriched_nbatches | prob_zero_enrichment | prob_zero_enrichment_rank | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| index | |||||||||||||||||||||
| WBGene00018727 | 1.0000 | 0.0000 | 18.420681 | 0.001506 | 0.000004 | 0.0 | 0.25 | 8.083410 | 8.553434 | 2.452872 | ... | -inf | WBGene00018727 | jbts-14 | Is involved in non-motile cilium assembly. Loc... | True | 0.923044 | 0.843094 | 1 | 0.1451 | 18051.0 |
| WBGene00004372 | 1.0000 | 0.0000 | 18.420681 | 0.000978 | 0.000005 | 0.0 | 0.25 | 7.747259 | 7.895372 | 1.346017 | ... | -inf | WBGene00004372 | rig-5 | Is an ortholog of human LSAMP (limbic system a... | True | 0.928574 | 0.814163 | 1 | 0.1740 | 18667.0 |
| WBGene00007266 | 1.0000 | 0.0000 | 18.420681 | 0.000556 | 0.000012 | 0.0 | 0.25 | 5.491581 | 5.608122 | 1.399333 | ... | -inf | WBGene00007266 | C03A3.1 | Is enriched in nervous system and pharynx base... | True | 0.714697 | 0.654044 | 1 | 0.2390 | 19470.0 |
| WBGene00007119 | 1.0000 | 0.0000 | 18.420681 | 0.000597 | 0.000009 | 0.0 | 0.25 | 5.797742 | 5.903028 | 1.622160 | ... | -inf | WBGene00007119 | calf-1 | Is involved in several processes, including de... | True | 0.832276 | 0.797743 | 1 | 0.1644 | 18475.0 |
| WBGene00043097 | 0.9998 | 0.0002 | 8.516543 | 0.000014 | 0.000153 | 0.0 | 0.25 | -3.516246 | -3.542899 | 0.972021 | ... | -3.698796 | WBGene00043097 | C02D5.4 | Is an ortholog of human GSTO1 (glutathione S-t... | True | 0.919165 | 0.881716 | 1 | 0.1124 | 17138.0 |
5 rows × 29 columns
de_change_importance = model.differential_expression(
idx1=cell_idx1, # we use the same cells as chosen before
idx2=cell_idx2,
weights="importance",
filter_outlier_cells=True,
batch_correction=True,
)
# manipulate the DE results for plotting
de_change_importance["log10_pscore"] = np.log10(de_change_importance["proba_not_de"])
de_change_importance = de_change_importance.join(adata.var, how="inner")
DE...: 100%|██████████| 1/1 [00:52<00:00, 52.94s/it]
de_comp = pd.concat(
[
de_change_importance.assign(flavor="importance"),
de_change_uniform.assign(flavor="uniform"),
]
)
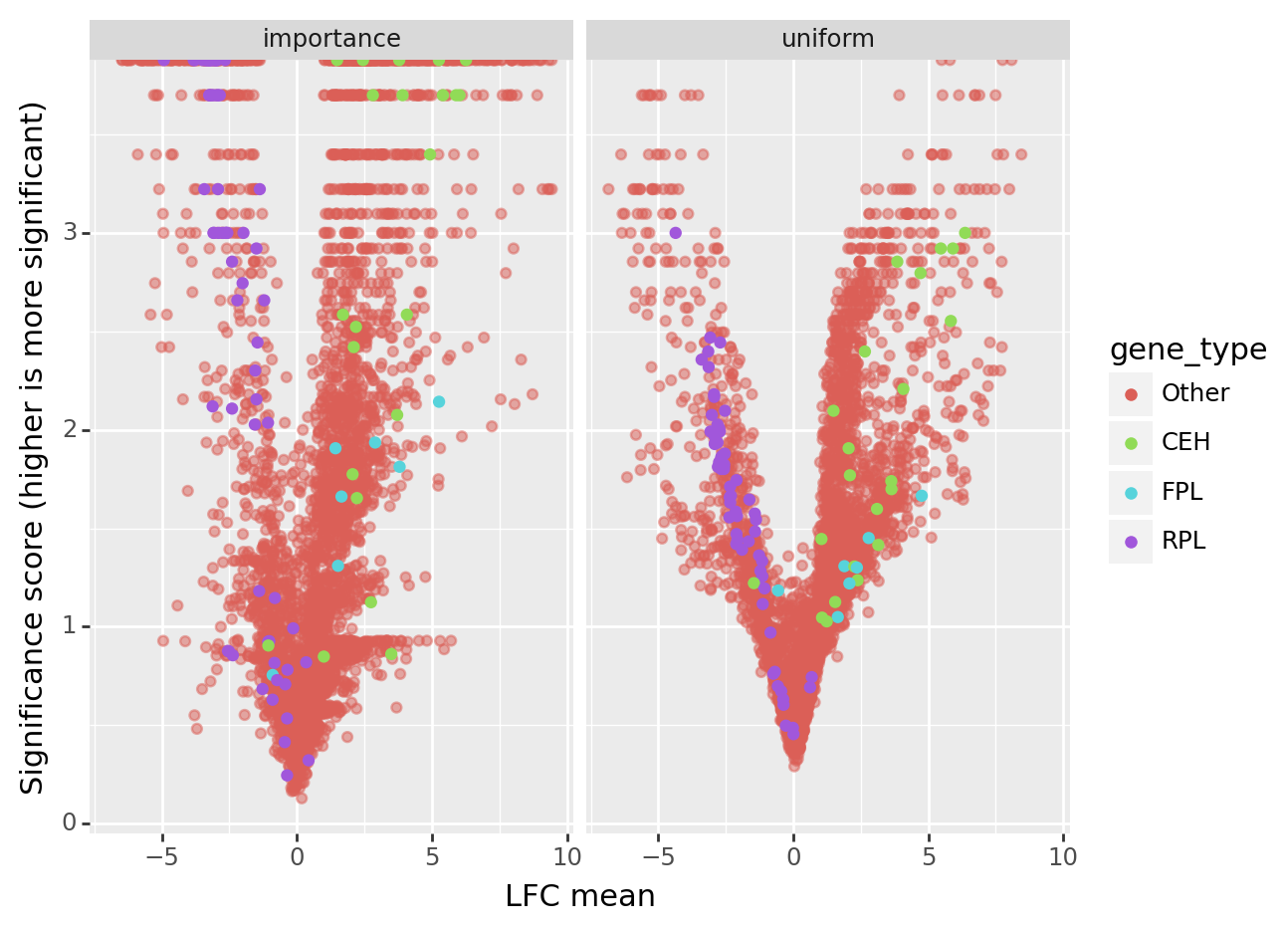
de_comp["gene_type"] = "Other"
de_comp.loc[lambda x: x["gene_name"].str.contains("rpl-"), "gene_type"] = "RPL"
de_comp.loc[lambda x: x["gene_name"].str.contains("ceh-"), "gene_type"] = "CEH"
de_comp.loc[lambda x: x["gene_name"].str.contains("flp-"), "gene_type"] = "FPL"
(
p9.ggplot(de_comp, p9.aes("lfc_mean", "-log10_pscore", color="gene_type"))
+ p9.geom_point(
de_comp.query("gene_type == 'Other'"), alpha=0.5
) # Plot other genes with transparence
+ p9.geom_point(de_comp.query("gene_type != 'Other'"))
+ p9.labs(x="LFC mean", y="Significance score (higher is more significant)")
+ p9.facet_wrap("flavor")
)
<Figure Size: (640 x 480)>
Contrary to the uniform flavor, the importance flavor returns sharper posterior probability scores (posterior probability near 0 or 1, respectively in case of equal expression or differential expression).
Heatmap of top expressed genes#
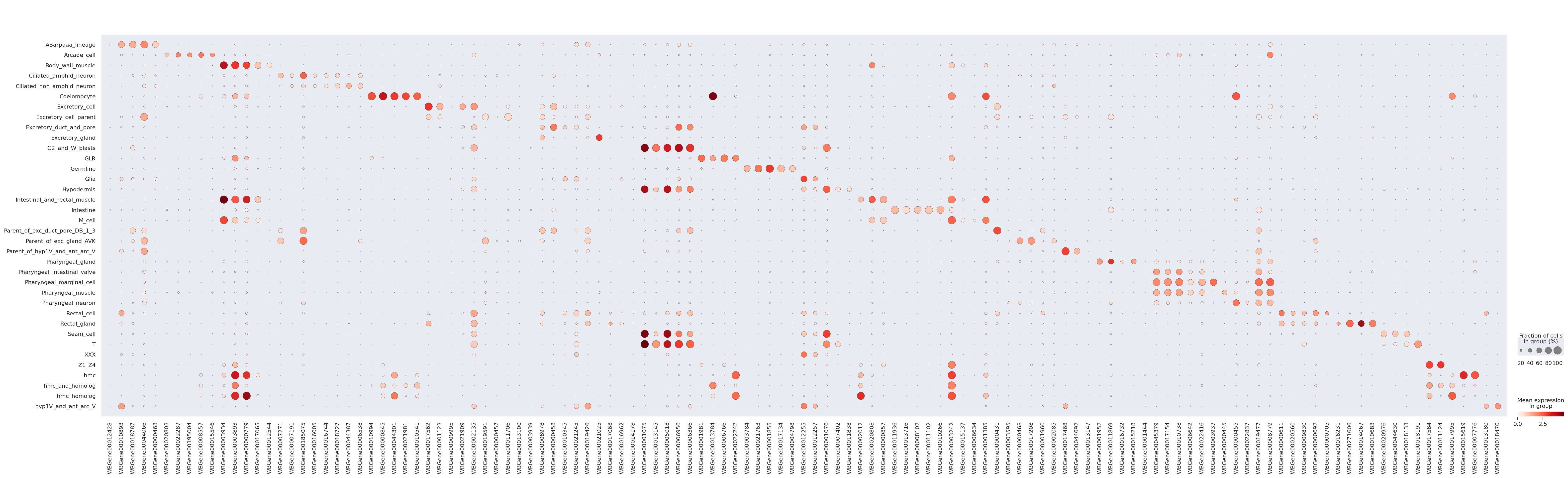
Now we perform DE between each cell type vs all other cells and make a heatmap of the result.
# here we do a 1-vs-all DE test, which compares each cell type with all others
# this returns the concatenation of all 1vsall results, contained in a DataFrame
change_per_cluster_de = model.differential_expression(groupby="cell_type")
DE...: 100%|██████████| 37/37 [02:21<00:00, 3.82s/it]
We focus on cell-types with at least 500 cells, and which have annotations to facilitate heatmap visualization
cell_types = (
adata.obs["cell_type"]
.value_counts()
# .loc[lambda x: (x >= 500) & (x.index != "nan")]
.loc[lambda x: x.index != "nan"]
.to_frame("n_cells")
)
cell_types.loc[:, "associated_test"] = cell_types.index.astype(str) + " vs Rest"
# This cell extracts list of top 5 upregulated genes for every cell-type
marker_genes = (
change_per_cluster_de.reset_index()
.loc[lambda x: x.comparison.isin(cell_types.associated_test.values)]
.groupby("comparison")
.apply(
lambda x: x.sort_values("lfc_mean", ascending=False).iloc[:5]
) # Select top 5 DE genes per comparison
.reset_index(drop=True)["index"]
.unique()
)
adata_log = adata[adata.obs.cell_type.isin(cell_types.index.values)].copy()
sc.pp.normalize_total(adata_log)
sc.pp.log1p(adata_log)
sc.pl.dotplot(adata_log, marker_genes, groupby="cell_type")